Perform a voxel-wise t-test
Edited/commented by Bogdan Petre on 9/14/2016, Tor Wager July 2018
In this example we perform a second level analysis on first level statistical parametric maps. Specifically we use a t-test to obtain an ordinary least squares estimate for the group level parameter.
The example uses the emotion regulation data provided with CANlab_Core_Tools. This dataset consists of a set of contrast images for 30 subjects from a first level analysis. The contrast is [reappraise neg vs. look neg], reappraisal of negative images vs. looking at matched negative images.
These data were published in: Wager, T. D., Davidson, M. L., Hughes, B. L., Lindquist, M. A., Ochsner, K. N.. (2008). Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron, 59, 1037-50.
By the end of this example we will have regenerated the results of figure 2A of this paper.
Contents
Dependencies:
% Matlab statistics toolbox % Matlab signal processing toolbox % Statistical Parametric Mapping (SPM) software https://www.fil.ion.ucl.ac.uk/spm/ % For full functionality, the full suite of CANlab toolboxes is recommended. See here: Installing Tools
Executive summary of the whole analysis
As a summary, here is a complete set of commands to load the data and run the entire analysis:
img_obj = load_image_set('emotionreg'); % Load a dataset
t = ttest(img_obj); % Do a group t-test
t = threshold(t, .05, 'fdr', 'k', 10); % Threshold with FDR q < .05 and extent threshold of 10 contiguous voxels% Show regions and print a table with labeled regions: montage(t); drawnow, snapnow; % Show results on a slice display r = region(t); % Turn t-map into a region object with one element per contig region table(r); % Print a table of results using new region names
Now, let's walk through it step by step.
Load sample data
% We can load this data using load_image_set(), which produces an fmri_data % object. Data loading exceeds the scope of this tutorial, but a more % indepth demosntration may be provided by load_a_sample_dataset.m [image_obj, networknames, imagenames] = load_image_set('emotionreg');
Loaded images: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii
Do a t-test
% Voxel-wise tests estimate parameters for each voxel independently. The % fmri_data/ttest function performs this just like the native matlab % ttest() function, and similarly returns a p-value and t-stat. Unlike the % matlab ttest() function, fmri_data/ttest performs this for every voxel in % the fmri_data object. All voxels are evaluated independently, but all are % evaluated nonetheless. A statistic_image is returned. t = ttest(image_obj);
One-sample t-test Calculating t-statistics and p-values
Visualize the results
There are many options. See methods(statistic_image) and methods(region)
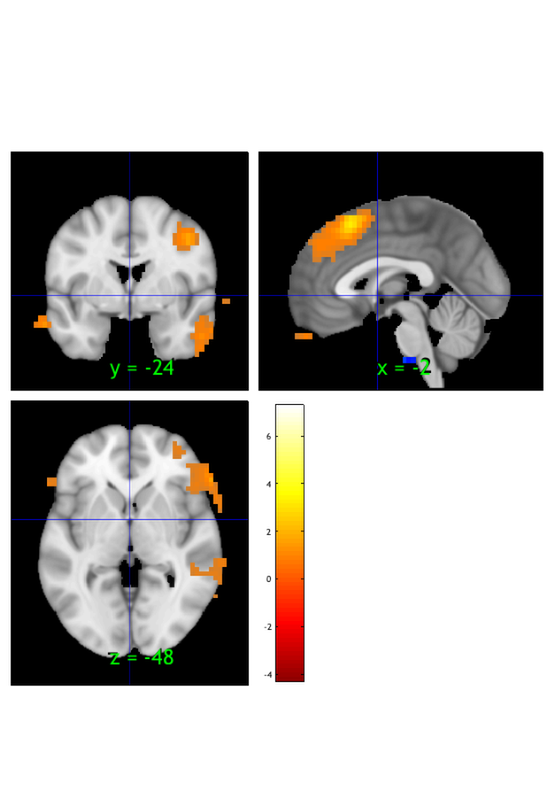
orthviews(t) drawnow, snapnow; % As can be seen, the ttest() result is unthresholded. The threshold % function can be used to apply a desired alpha level using any of a number % of methods. Here FDR is used to control for alpha=0.05. Note that no % information is erased when performing thresholding on a statistic_image. t = threshold(t, .05, 'fdr'); orthviews(t) drawnow, snapnow; % Many neuroimaging packages (e.g., SPM and FSL) do one-tailed tests % (with one-tailed p-values) and only show you positive effects % (i.e., relative activations, not relative deactivations). % All the CANlab tools do two-sided tests, report two-tailed p-values. % By default, hot colors (orange/yellow) will show activations, and cool % colors (blues) show deactivations. % We can also apply an "extent threshold" of > 10 contiguous voxels: t = threshold(t, .05, 'fdr', 'k', 10); orthviews(t) drawnow, snapnow; % montage is another visualization method. This function may require a % relatively large amount of memory, depending on the resolution of the % anatomical underlay image you use. We recommend around 8GB of free memory. % create_figure('montage'); axis off montage(t) drawnow, snapnow;
SPM12: spm_check_registration (v6245) 23:49:08 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
ans =
1×1 cell array
{1×1 struct}
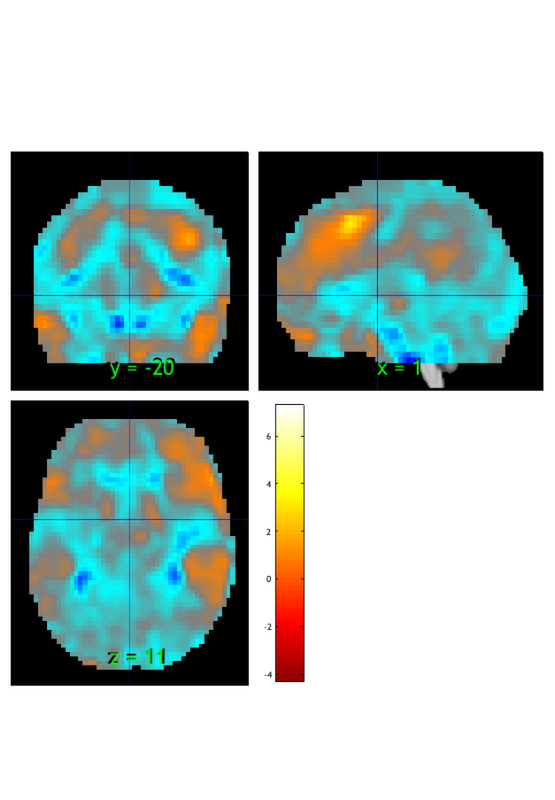
Image 1 FDR q < 0.050 threshold is 0.002549
Image 1
27 contig. clusters, sizes 1 to 1472
Positive effect: 2502 voxels, min p-value: 0.00000000
Negative effect: 37 voxels, min p-value: 0.00022793
SPM12: spm_check_registration (v6245) 23:49:11 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
ans =
1×1 cell array
{1×27 struct}
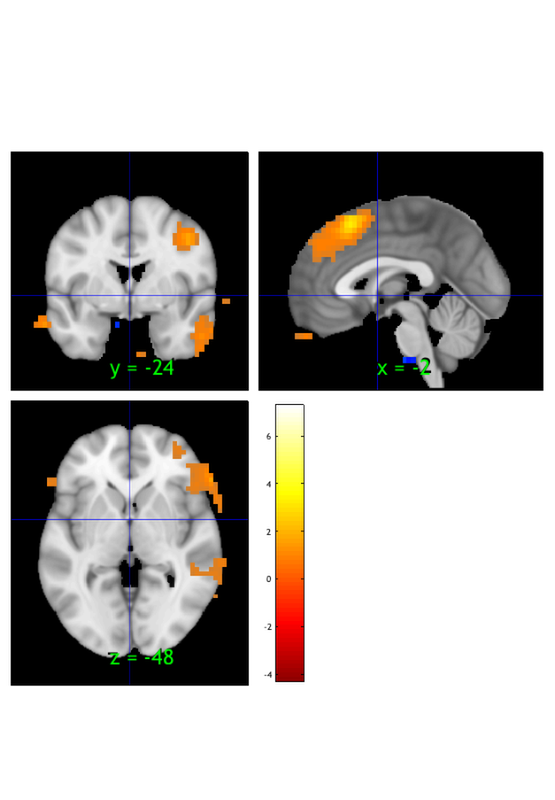
Image 1 FDR q < 0.050 threshold is 0.002549
Image 1
9 contig. clusters, sizes 10 to 1472
Positive effect: 2460 voxels, min p-value: 0.00000000
Negative effect: 32 voxels, min p-value: 0.00022793
SPM12: spm_check_registration (v6245) 23:49:13 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
ans =
1×1 cell array
{1×9 struct}
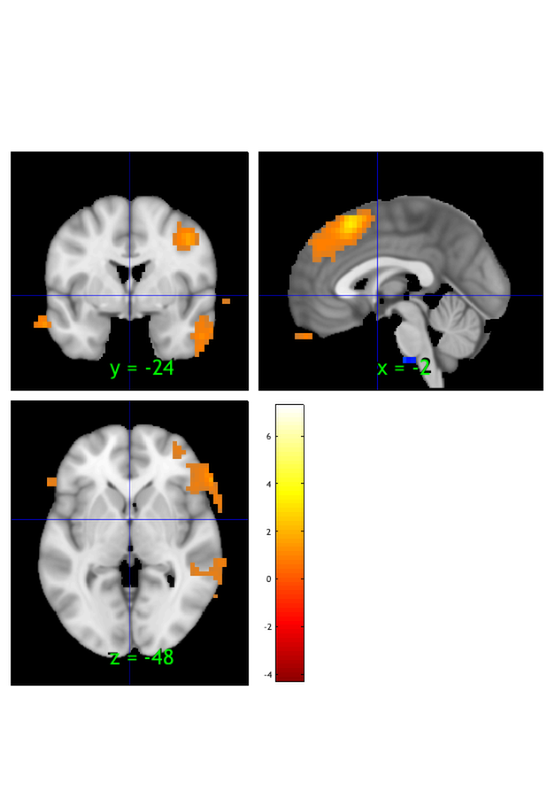
Setting up fmridisplay objects
This takes a lot of memory, and can hang if you have too little.
Grouping contiguous voxels: 9 regions
sagittal montage: 31 voxels displayed, 2461 not displayed on these slices
sagittal montage: 97 voxels displayed, 2395 not displayed on these slices
sagittal montage: 76 voxels displayed, 2416 not displayed on these slices
axial montage: 808 voxels displayed, 1684 not displayed on these slices
axial montage: 904 voxels displayed, 1588 not displayed on these slices
ans =
fmridisplay with properties:
overlay: '/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img'
SPACE: [1×1 struct]
activation_maps: {[1×1 struct]}
montage: {1×5 cell}
surface: {}
orthviews: {}
history: {}
history_descrip: []
additional_info: ''
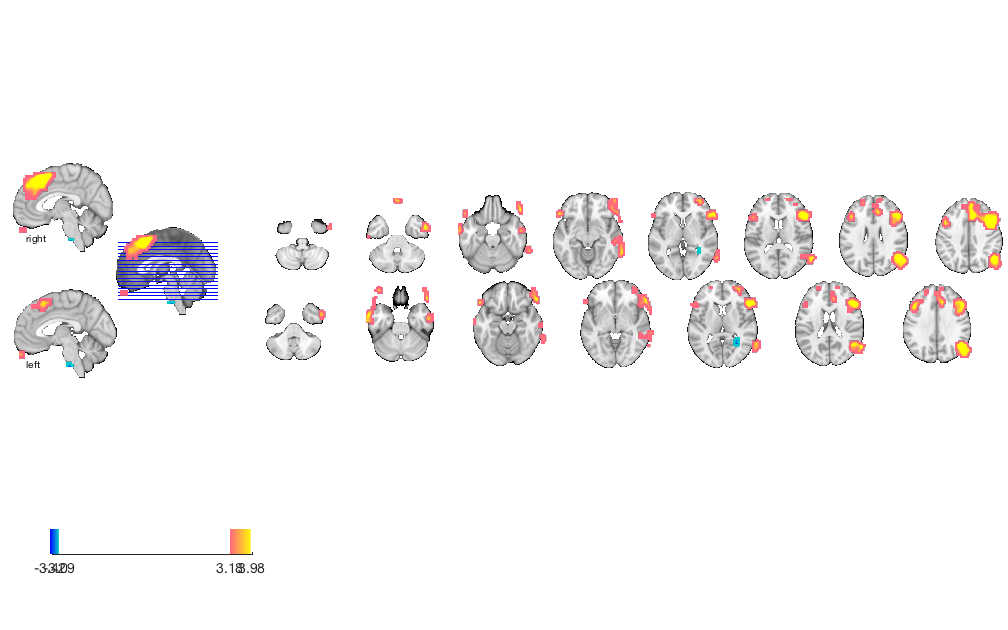
Print a table of results
% First, we'll have to convert to another object type, a "region object". % This object groups voxels together into "blobs" (often of contiguous % voxels). It does many things that other object types do, and inter-operates % with them closely. See methods(region) for more info. % Create a region object called "r", with contiguous blobs from the % thresholded t-map: r = region(t); % Print the table: table(r); % You can get help and options for any object method, like "table". But % because the method names are simple and often overlap with other Matlab functions % and toolboxes (this is OK for objects!), you will often want to specify % the object type as well, as follows: help region.table
Grouping contiguous voxels: 9 regions
____________________________________________________________________________________________________________________________________________
Positive Effects
Region Volume XYZ maxZ modal_label_descriptions Perc_covered_by_label Atlas_regions_covered region_index
__________________ __________ _________________ ______ ________________________ _____________________ _____________________ ____________
'Ctx_TE1a_R' 9864 55 0 -27 3.9997 'Cortex_Default_ModeA' 30 2 1
'Multiple regions' 57152 55 -58 23 4.7899 'Cortex_Default_ModeA' 7 10 6
'Multiple regions' 31664 -55 21 9 4.1824 'Cortex_Default_ModeB' 9 10 2
'Multiple regions' 1.4038e+05 31 28 36 7.0345 'Cortex_Default_ModeB' 4 38 4
'Ctx_9a_L' 4336 -24 52 27 3.443 'Cortex_Default_ModeB' 35 1 7
'Ctx_10v_L' 3752 -3 58 -32 3.943 'Cortex_Limbic' 7 0 3
'No label' 912 -45 52 -23 3.8446 'No description' 0 0 5
Negative Effects
Region Volume XYZ maxZ modal_label_descriptions Perc_covered_by_label Atlas_regions_covered region_index
______________ ______ ________________ _______ __________________________ _____________________ _____________________ ____________
'Bstem_Pons_L' 2016 -3 -24 -50 -3.6859 'Brainstem' 22 0 8
'Ctx_ProS_R' 2832 28 -48 9 -3.3405 'Cortex_Visual_Peripheral' 12 0 9
____________________________________________________________________________________________________________________________________________
Regions labeled by reference atlas CANlab_2018_combined
Volume: Volume of contiguous region in cubic mm.
MaxZ: Signed max over Z-score for each voxel.
Atlas_regions_covered: Number of reference atlas regions covered at least 25% by the region. This relates to whether the region covers
multiple reference atlas regions
Region: Best reference atlas label, defined as reference region with highest number of in-region voxels. Regions covering >25% of >5
regions labeled as "Multiple regions"
Perc_covered_by_label: Percentage of the region covered by the label.
Ref_region_perc: Percentage of the label region within the target region.
modal_atlas_index: Index number of label region in reference atlas
all_regions_covered: All regions covered >5% in descending order of importance
For example, if a region is labeled 'TE1a' and Perc_covered_by_label = 8, Ref_region_perc = 38, and Atlas_regions_covered = 17, this means
that 8% of the region's voxels are labeled TE1a, which is the highest percentage among reference label regions. 38% of the region TE1a is
covered by the region. However, the region covers at least 25% of 17 distinct labeled reference regions.
References for atlases:
Beliveau, Vincent, Claus Svarer, Vibe G. Frokjaer, Gitte M. Knudsen, Douglas N. Greve, and Patrick M. Fisher. 2015. “Functional
Connectivity of the Dorsal and Median Raphe Nuclei at Rest.” NeuroImage 116 (August): 187–95.
Bär, Karl-Jürgen, Feliberto de la Cruz, Andy Schumann, Stefanie Koehler, Heinrich Sauer, Hugo Critchley, and Gerd Wagner. 2016. ?Functional
Connectivity and Network Analysis of Midbrain and Brainstem Nuclei.? NeuroImage 134 (July):53?63.
Diedrichsen, Jörn, Joshua H. Balsters, Jonathan Flavell, Emma Cussans, and Narender Ramnani. 2009. A Probabilistic MR Atlas of the Human
Cerebellum. NeuroImage 46 (1): 39?46.
Fairhurst, Merle, Katja Wiech, Paul Dunckley, and Irene Tracey. 2007. ?Anticipatory Brainstem Activity Predicts Neural Processing of Pain
in Humans.? Pain 128 (1-2):101?10.
Fan 2016 Cerebral Cortex; doi:10.1093/cercor/bhw157
Glasser, Matthew F., Timothy S. Coalson, Emma C. Robinson, Carl D. Hacker, John Harwell, Essa Yacoub, Kamil Ugurbil, et al. 2016. A
Multi-Modal Parcellation of Human Cerebral Cortex. Nature 536 (7615): 171?78.
Keren, Noam I., Carl T. Lozar, Kelly C. Harris, Paul S. Morgan, and Mark A. Eckert. 2009. “In Vivo Mapping of the Human Locus Coeruleus.”
NeuroImage 47 (4): 1261–67.
Keuken, M. C., P-L Bazin, L. Crown, J. Hootsmans, A. Laufer, C. Müller-Axt, R. Sier, et al. 2014. “Quantifying Inter-Individual Anatomical
Variability in the Subcortex Using 7 T Structural MRI.” NeuroImage 94 (July): 40–46.
Krauth A, Blanc R, Poveda A, Jeanmonod D, Morel A, Székely G. (2010) A mean three-dimensional atlas of the human thalamus: generation from
multiple histological data. Neuroimage. 2010 Feb 1;49(3):2053-62. Jakab A, Blanc R, Berényi EL, Székely G. (2012) Generation of
Individualized Thalamus Target Maps by Using Statistical Shape Models and Thalamocortical Tractography. AJNR Am J Neuroradiol. 33:
2110-2116, doi: 10.3174/ajnr.A3140
Nash, Paul G., Vaughan G. Macefield, Iven J. Klineberg, Greg M. Murray, and Luke A. Henderson. 2009. ?Differential Activation of the Human
Trigeminal Nuclear Complex by Noxious and Non-Noxious Orofacial Stimulation.? Human Brain Mapping 30 (11):3772?82.
Pauli 2018 Bioarxiv: CIT168 from Human Connectome Project data
Pauli, Wolfgang M., Amanda N. Nili, and J. Michael Tyszka. 2018. ?A High-Resolution Probabilistic in Vivo Atlas of Human Subcortical Brain
Nuclei.? Scientific Data 5 (April): 180063.
Pauli, Wolfgang M., Randall C. O?Reilly, Tal Yarkoni, and Tor D. Wager. 2016. ?Regional Specialization within the Human Striatum for
Diverse Psychological Functions.? Proceedings of the National Academy of Sciences of the United States of America 113 (7): 1907?12.
Sclocco, Roberta, Florian Beissner, Gaelle Desbordes, Jonathan R. Polimeni, Lawrence L. Wald, Norman W. Kettner, Jieun Kim, et al. 2016.
?Neuroimaging Brainstem Circuitry Supporting Cardiovagal Response to Pain: A Combined Heart Rate Variability/ultrahigh-Field (7 T)
Functional Magnetic Resonance Imaging Study.? Philosophical Transactions. Series A, Mathematical, Physical, and Engineering Sciences 374
(2067). rsta.royalsocietypublishing.org. https://doi.org/10.1098/rsta.2015.0189.
Shen, X., F. Tokoglu, X. Papademetris, and R. T. Constable. 2013. “Groupwise Whole-Brain Parcellation from Resting-State fMRI Data for
Network Node Identification.” NeuroImage 82 (November): 403–15.
Zambreanu, L., R. G. Wise, J. C. W. Brooks, G. D. Iannetti, and I. Tracey. 2005. ?A Role for the Brainstem in Central Sensitisation in
Humans. Evidence from Functional Magnetic Resonance Imaging.? Pain 114 (3):397?407.
Note: Region object r(i).title contains full list of reference atlas regions covered by each cluster.
____________________________________________________________________________________________________________________________________________
Print a table of all regions in a region object (cl). Return labeled
clusters (separated by those with positive and negative peak values) and
a Matlab table object with the relevant information.
:Usage:
::
[poscl, negcl] = table(cl, [optional inputs])
:Optional inputs:
**k:**
Print only regions with k or more contiguous voxels
**nosep:**
do not separate cl with pos and neg effects based on peak in .val
**names:**
name clusters manually before printing to table and output; saves in .shorttitle field
**forcenames:**
force manual naming of cl by removing existing names in .shorttitle field
:Outputs:
Returns region objects for cl with pos and neg effects
- autolabeled if Neuroimaging_Pattern_Masks and atlas tools are available on Matlab path
- limited by size if entered
- manually named if requested (see optional inputs)
..
Author and copyright information:
Copyright (C) 2011 Tor Wager
This program is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
You should have received a copy of the GNU General Public License
along with this program. If not, see <http://www.gnu.org/licenses/>.
..
:Examples:
-------------------------------------------------------------------------
::
Example 1:
% Complete group analysis of a standard dataset
% Do analysis and prep results region object:
img_obj = load_image_set('emotionreg'); % Load a dataset
t = ttest(img_obj, .005, 'unc'); % Do a group t-test
t = threshold(t, .005, 'unc', 'k', 10); % Re-threshold with extent threshold of 10 contiguous voxels
r = region(t); % Turn t-map into a region object with one element per contig region
Label regions and print a table:
[r, region_table, table_legend_text] = autolabel_regions_using_atlas(r);
% Label regions. Can be skipped because 'table' below attempts to do this automatically
table(r); % Print a table of results using new region names
[rpos, rneg] = table(r); % Print and table and return region object separated into regions with positive vs. negative statistic values (from .Z field)
Write the t-map to disk
% Now we need to save our results. You can save the objects in your % workspace or you can write your resulting thresholded map to an analyze % file. The latter may be useful for generating surface projections using % Caret or FreeSurfer for instance. % % Thresholding did not actually eliminate nonsignificant voxels from our % statistic_image object (t). If we simply write out that object, we will % get t-statistics for all voxels. t.fullpath = fullfile(pwd, 'example_t_image.nii'); write(t) % If we use the 'thresh' option, we'll write thresholded values: write(t, 'thresh') t_reloaded = statistic_image(t.fullpath, 'type', 'generic'); orthviews(t_reloaded)
Writing:
/Users/torwager/Documents/GitHub/Test_mediation/example_t_image.nii
Writing thresholded statistic image.
Writing:
/Users/torwager/Documents/GitHub/Test_mediation/example_t_image.nii
SPM12: spm_check_registration (v6245) 23:49:26 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
Displaying image 1, 2492 voxels: example_t_image.nii
ans =
1×1 cell array
{1×9 struct}