Contents
- HOW TO WRITE FMRI_DATA TO IMAGE FILE ON DISK
- 1) load sample mask
- grab one image from those 7
- write to file
- read back in and view to check its correct
- 2) load sample statistical image
- threshold at an arbitrary threshold. This will create a statistic_image
- write to file
- read back in and view to check its correct
Dependencies:
% Matlab statistics toolbox % Matlab signal processing toolbox % Statistical Parametric Mapping (SPM) software https://www.fil.ion.ucl.ac.uk/spm/ % For full functionality, the full suite of CANlab toolboxes is recommended. See here: Installing Tools
HOW TO WRITE FMRI_DATA TO IMAGE FILE ON DISK
% canlabcore fmri data objects can be easily written to standard image file % formats. The main function for this is write() % this walkthrough shows how to do this for 1) a mask image, and 2) a % statistic image
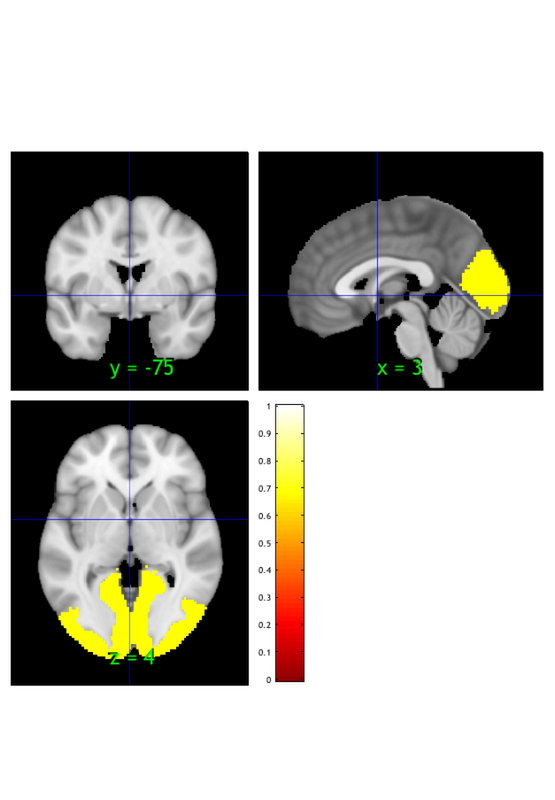
1) load sample mask
You will need the Neuroimaging_Pattern_Masks repository installed to find these images.
dat = load_image_set('bucknerlab'); % loads 7 masks from Yeo et al.
Loaded images: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Combined_multiatlas_ROI_masks/rBucknerlab_7clusters_SPMAnat_Other_combined.img
grab one image from those 7
oneimg = dat.get_wh_image(1);
write to file
see help for write() for more options
fname = 'dummy.nii'; % outname file name. the extension (.nii, .img) determines the format. write(oneimg, 'fname', fname);
Writing: dummy.nii
read back in and view to check its correct
orthviews(fmri_data(fname))
Using default mask: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/brainmask.nii
loading mask. mapping volumes.
checking that dimensions and voxel sizes of volumes are the same.
Pre-allocating data array. Needed: 1409312 bytes
Loading image number: 1
Elapsed time is 0.020971 seconds.
Image names entered, but fullpath attribute is empty. Getting path info.
SPM12: spm_check_registration (v6245) 19:30:11 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
Grouping contiguous voxels: 3 regions
ans =
1×1 cell array
{1×3 region}
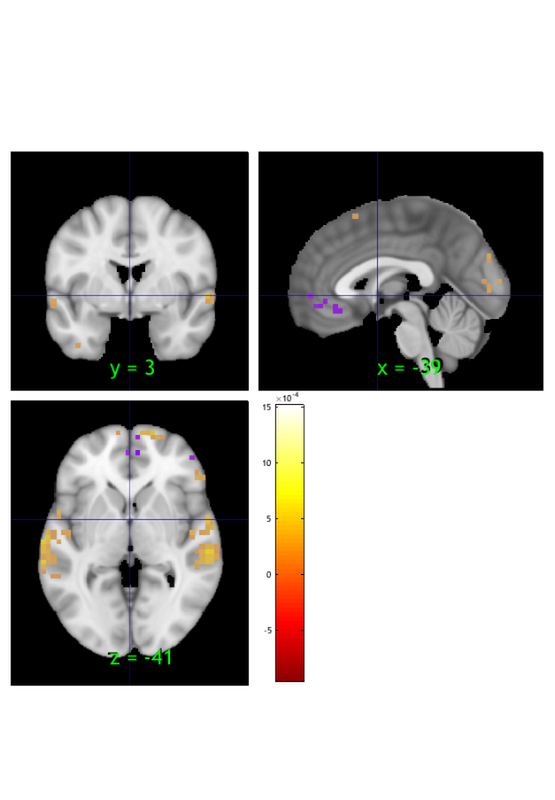
2) load sample statistical image
dat = load_image_set('kragelemotion'); % loads 7 masks from Kragel et al. % grab one image from those 7 oneimg = dat.get_wh_image(1);
Loaded images: /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_amused_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_angry_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_content_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_fearful_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_neutral_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_sad_group_emotion_PLS_beta_BSz_10000it.img /Users/torwager/Documents/GitHub/MasksPrivate/Masks_private/2015_Kragel_emotionClassificationBPLS/mean_3comp_surprised_group_emotion_PLS_beta_BSz_10000it.img
threshold at an arbitrary threshold. This will create a statistic_image
% keep values less than -.0005 or greater than .0005 si = threshold(oneimg, [-.0005 .0005], 'raw-outside'); % check that it looks fine orthviews(si);
Keeping vals outside of -0.001 to 0.001: 474 elements remain
SPM12: spm_check_registration (v6245) 19:30:14 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
Grouping contiguous voxels: 83 regions
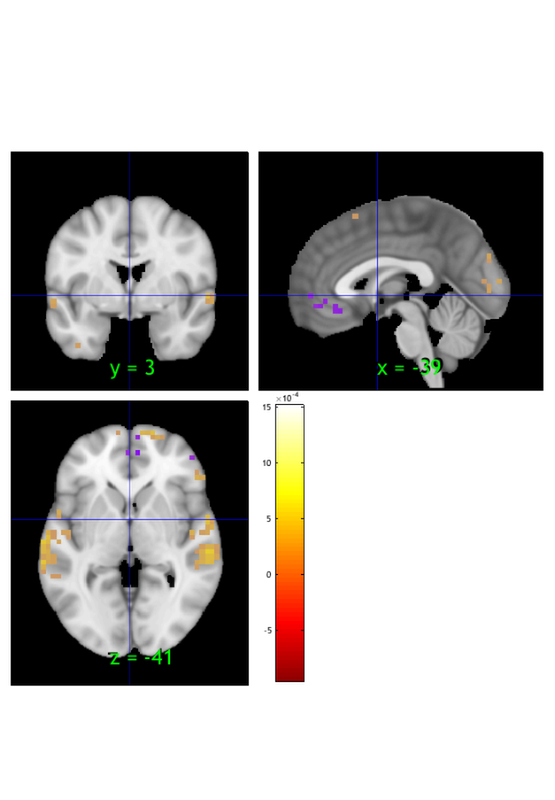
write to file
fname = 'dummy.nii'; % outname file name. the extension (.nii, .img) determines the format. write(si, 'fname', fname, 'thresh') % will write the *thresholded* image
Warning: Thresholding only works with statistic_image objects. Writing: dummy.nii
read back in and view to check its correct
orthviews(fmri_data(fname))
Using default mask: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/brainmask.nii
loading mask. mapping volumes.
checking that dimensions and voxel sizes of volumes are the same.
Pre-allocating data array. Needed: 188152 bytes
Loading image number: 1
Elapsed time is 0.008781 seconds.
Image names entered, but fullpath attribute is empty. Getting path info.
SPM12: spm_check_registration (v6245) 19:30:17 - 02/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
Grouping contiguous voxels: 83 regions
ans =
1×1 cell array
{1×83 region}