Installing CANlab Core Tools
Contents
About the CANlab Core Tools repository and CANlab toolboxes
The CANlab Core Tools repository is on https://github.com/canlab/CanlabCore It contains core tools for MRI/fMRI/PET analysis from the Cognitive and Affective Neuorscience Lab (Tor Wager, PI) and our collaborators. Many of these functions are needed to run other toolboxes, e.g., the CAN lab?s multilevel mediation and Martin Lindquist?s hemodynamic response estimation toolboxes.
The tools include object-oriented tools for doing neuroimaging analysis with simple commands and scripts that provide high-level functionality for neuroimaging analysis. For example, there is an "fmri_data" object type that contains neuroimaging datasets (both PET and fMRI data are ok, despite the name). If you have created and object called my_fmri_data_obj, then plot(my_fmri_data_obj) will generate a series of plots specific to neuroimaging data, including an interactive brain viewer (courtesy of SPM software). predict(my_fmri_data_obj) will perform cross-validated multivariate prediction of outcomes based on brain data. ica(my_fmri_data_obj) will perform independent components analysis on the data, and so forth.
There are a wide range of functions and techniques that we've used in the CANlab. Most of these are supported by specific functionality in our object-oriented tools. You can download other toolboxes from: https://github.com/canlab
And you can find an overview of core object-oriented tool functionality and demos/walkthroughs on https://canlab.github.io
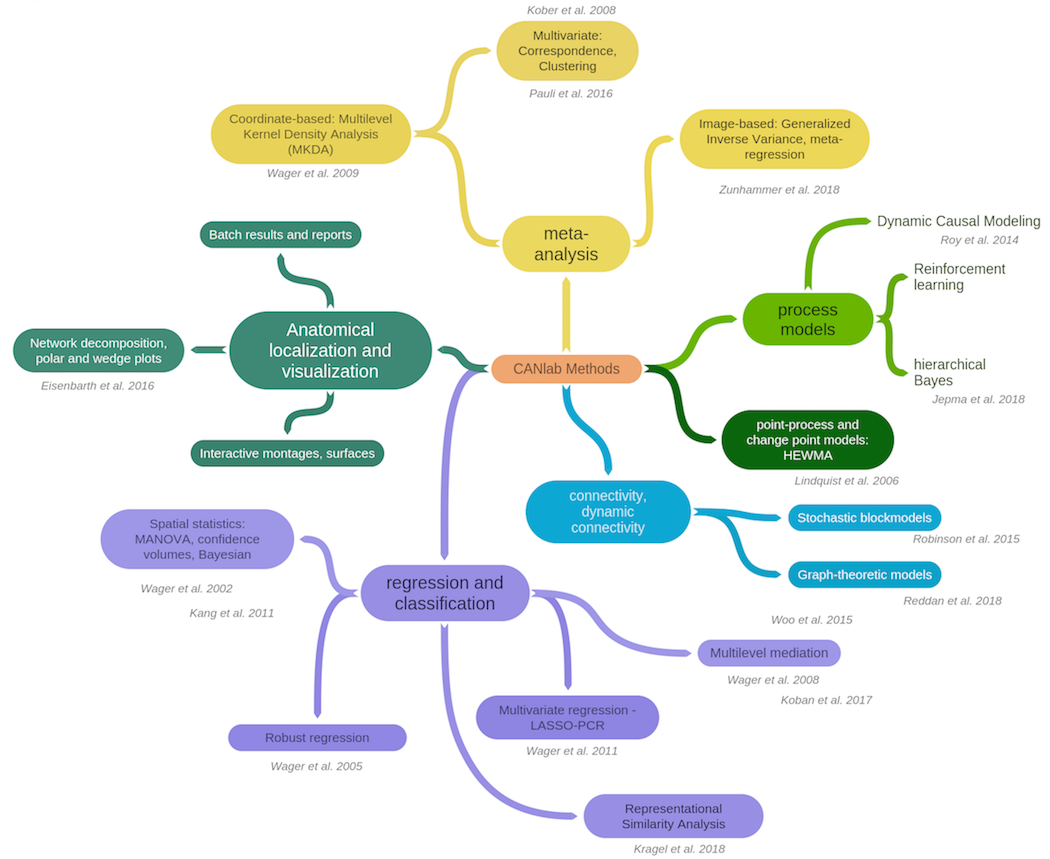
CANlab Toolboxes
Tutorials, overview, and help: https://canlab.github.io
Toolboxes and image repositories on github: https://github.com/canlab
| CANlab Core Tools | https://github.com/canlab/CanlabCore | CANlab Neuroimaging_Pattern_Masks repository | https://github.com/canlab/Neuroimaging_Pattern_Masks | CANlab_help_examples | https://github.com/canlab/CANlab_help_examples | M3 Multilevel mediation toolbox | https://github.com/canlab/MediationToolbox | M3 CANlab robust regression toolbox | https://github.com/canlab/RobustToolbox | M3 MKDA coordinate-based meta-analysis toolbox | https://github.com/canlab/Canlab_MKDA_MetaAnalysis |
Here are some other useful CANlab-associated resources:
| Paradigms_Public - CANlab experimental paradigms | https://github.com/canlab/Paradigms_Public | FMRI_simulations - brain movies, effect size/power | https://github.com/canlab/FMRI_simulations | CANlab_data_public - Published datasets | https://github.com/canlab/CANlab_data_public | M3 Neurosynth: Tal Yarkoni | https://github.com/neurosynth/neurosynth | M3 DCC - Martin Lindquist's dynamic correlation tbx | https://github.com/canlab/Lindquist_Dynamic_Correlation | M3 CanlabScripts - in-lab Matlab/python/bash | https://github.com/canlab/CanlabScripts |
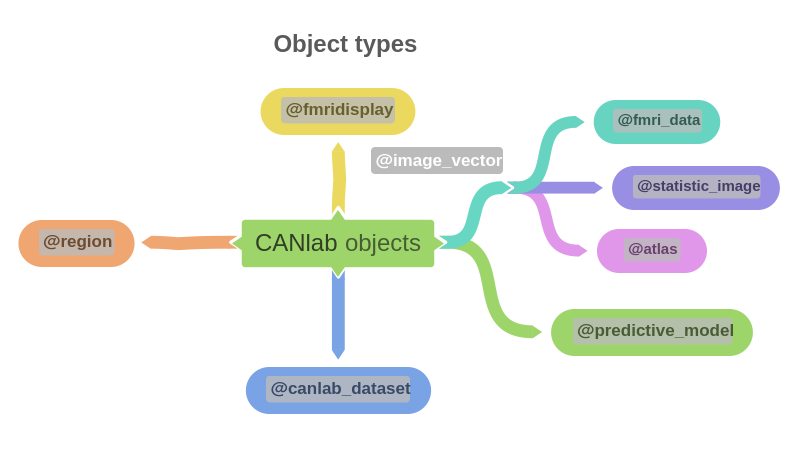
Object-oriented, interactive approach The core basis for interacting with CANlab tools is through object-oriented framework. A simple set of neuroimaging data-specific objects (or classes) allows you to perform interactive analysis using simple commands (called methods) that operate on objects.
Map of core object classes:
Dependencies
- Matlab statistics toolbox
- Matlab signal processing toolbox
- Statistical Parametric Mapping (SPM) software https://www.fil.ion.ucl.ac.uk/spm/ (this is used for image reading/writing and the orthviews function.)
For full functionality, the other toolboxes below are recommended:
Installing SPM
! Important: You will also need to install spm12 separately, and add it to your matlab path. The latest version at the time of writing is SPM12, which can be downloaded here:
https://www.fil.ion.ucl.ac.uk/spm/software/spm12/
Quick start instructions
- Download SPM12 from https://www.fil.ion.ucl.ac.uk/spm/
- Find the SPM12 folder and drag it into your command window. Matlab will go to that directory (execute the cd command).
- In Matlab, type addpath pwd. This adds the main SPM12 folder to your path.
- Type addpath canonical. This adds the canonical subfolder to your path.
- Go to https://github.com/canlab/ and click on CanlabCore
- Sign into Github (top right corner)
- Click "Clone or Download"
- Click "Open in Desktop" (if you have Github Desktop) to clone the repository
- Find the folder and drag it into your Matlab command window.
- In Matlab, type g = genpath(pwd). This lists all the subfolders in a string g
- Type addpath(g). This adds them to your path.
- Type savepath. This saves your path for future use.
Repeat the Clone/genpath/addpath steps for the Neuroimaging_Pattern_Masks repository, and save the path again.
Quick start: Batch install script
The main goal of the code below is to run: canlab_toolbox_setup This is a script that attempts to: # Check for CANlab repositories on your computer # Download any that are missing # Add them to your Matlab path with subfolders.
Note: Unfortunately as of Jan 2020 this does not work well on everyone's computers, due to OS-related variability. The workaround is to install them manually as per the instructions above.
canlab_toolbox_setup is in the CanlabCore repository. So before you run it the CANlab_Core_Tools must be added to your path with subfolders. Otherwise, you will get errors.
The script canlab_toolbox_setup can help download and install the toolboxes you need.
First, go to a folder where you want to install toolboxes/repositories on your hard drive. Mine are in "Github" and I've already installed CanlabCore, so let's find it and go there:
% Locate the CanlabCore Github files on your local computer: % ------------------------------------------------------------------------ mypath = what(fullfile('CanlabCore', 'CanlabCore')); % Check if we've got it if isempty(mypath) disp('Download CanlabCore from Github, and go to that folder in Matlab') disp('by dragging and dropping it from Finder or Explorer into the Matlab Command Window') return else % Add CanlabCore to Matlab path with subfolders g = genpath(mypath(1).path); addpath(g); end mypath = mypath(1).path; % Set the folder in which the repositories will be installed % ------------------------------------------------------------------------ % This is the location (folder) in which the repositories will be installed: base_dir_for_repositories = fileparts(fileparts(mypath)); cd(base_dir_for_repositories) fprintf('\nInstalling repositories in %s\n', base_dir_for_repositories); % Find the setup file, canlab_toolbox_setup.m % ------------------------------------------------------------------------ % This is the file setupfile = fullfile(mypath, 'canlab_toolbox_setup.m'); % Make sure we've got it if ~exist(setupfile, 'file') disp('Something went wrong. I can''t find canlab_toolbox_setup.m'); disp('This file should be included in the CanlabCore Github repository.') end % Run the setup script: % ------------------------------------------------------------------------ % % Note: This can be system-dependent and may not work on all computers. % If it doesn't, you can default to cloning the CANLab repositories and % adding them with subfolders to your Matlab path. The most important ones % starting out are CanlabCore and Neuroimaging_Pattern_Masks % e.g., % |git clone https://github.com/canlab/CanlabCore.git| % |git clone https://github.com/canlab/Neuroimaging_Pattern_Masks.git| canlab_toolbox_setup % This will attempt to locate toolboxes, add them to your path, and give % you the option to download them from Github if it can't find them. % It looks for and installs toolboxes in the current directory, % which (thanks to the code above) is the path name in base_dir_for_repositories
Installing repositories in /Users/torwager/Documents/GitHub ----------------------------------------- Checking and/or installing CANlab tools in: /Users/torwager/Documents/GitHub ----------------------------------------- Looking for CANlab repositories and adding to Matlab path: CANlab core tools - Object-oriented interactive analysis and tools used in other functions CanlabCore CANlab help examples - Example scripts and html output, second-level analysis batch scripts CANlab_help_examples MKDA meta-analysis toolbox - Coordinate-based meta-analysis of neuroimaging data Canlab_MKDA_MetaAnalysis FMRI_simulations - Misc tools for simulating and testing power and image data FMRI_simulations Private masks and signatures - Selected models shared on request MasksPrivate M3 Mediation toolbox - Single and Multi-level mediation tools MediationToolbox Neuroimaging_Pattern_Masks - meta-analysis masks, signatures, parcellations Neuroimaging_Pattern_Masks Robust regression toolbox - Voxel-wise robust regression for neuroimaging data RobustToolbox CanlabPrivate - Private repository for lab members and collaborators CanlabPrivate ----------------------------------------- ----------------------------------------- Repository: CANlab core tools - Object-oriented interactive analysis and tools used in other functions Site:CanlabCore Finding repo: Running command: find /Users/torwager/Documents/GitHub/CanlabCore -name "fmri_data.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/CanlabCore ----------------------------------------- Repository: CANlab help examples - Example scripts and html output, second-level analysis batch scripts Site:CANlab_help_examples Finding repo: Running command: find /Users/torwager/Documents/GitHub/CANlab_help_examples -name "a2_second_level_toolbox_check_dependencies.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/CANlab_help_examples ----------------------------------------- Repository: MKDA meta-analysis toolbox - Coordinate-based meta-analysis of neuroimaging data Site:Canlab_MKDA_MetaAnalysis Finding repo: Running command: find /Users/torwager/Documents/GitHub/Canlab_MKDA_MetaAnalysis -name "i_density.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/Canlab_MKDA_MetaAnalysis ----------------------------------------- Repository: FMRI_simulations - Misc tools for simulating and testing power and image data Site:FMRI_simulations Finding repo: Running command: find /Users/torwager/Documents/GitHub/FMRI_simulations -name "power_figure3_num_comparisons.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/FMRI_simulations ----------------------------------------- Repository: Private masks and signatures - Selected models shared on request Site:MasksPrivate Finding repo: Running command: find /Users/torwager/Documents/GitHub/MasksPrivate -name "apply_nps.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/MasksPrivate ----------------------------------------- Repository: M3 Mediation toolbox - Single and Multi-level mediation tools Site:MediationToolbox Finding repo: Running command: find /Users/torwager/Documents/GitHub/MediationToolbox -name "mediation_brain.m" Warning: Function vecnorm has the same name as a MATLAB builtin. We suggest you rename the function to avoid a potential name conflict. Found and added to path with subfolders: /Users/torwager/Documents/GitHub/MediationToolbox ----------------------------------------- Repository: Neuroimaging_Pattern_Masks - meta-analysis masks, signatures, parcellations Site:Neuroimaging_Pattern_Masks Finding repo: Running command: find /Users/torwager/Documents/GitHub/Neuroimaging_Pattern_Masks -name "apply_all_signatures.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/Neuroimaging_Pattern_Masks ----------------------------------------- Repository: Robust regression toolbox - Voxel-wise robust regression for neuroimaging data Site:RobustToolbox Finding repo: Running command: find /Users/torwager/Documents/GitHub/RobustToolbox -name "robust_results_batch.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/RobustToolbox ----------------------------------------- Repository: CanlabPrivate - Private repository for lab members and collaborators Site:CanlabPrivate Finding repo: Running command: find /Users/torwager/Documents/GitHub/CanlabPrivate -name "power_calc.m" Found and added to path with subfolders: /Users/torwager/Documents/GitHub/CanlabPrivate ----------------------------------------- All CANlab repositories previously installed. Saving path