Contents
- Load dataset with images and print descriptives
- Collect some variables we need
- Plot the ratings
- Prediction
- Run the Base model
- Plot the results
- Summarize within-person classification accuracy
- Visualize weight map
- Bootstrap weights: Get most reliable weights and p-values for voxels
- Try normalizing/scaling data
- Try selecting an a priori 'network of interest'
- re-run prediction and plot
- Other ideas
Dependencies:
% Matlab statistics toolbox % Matlab signal processing toolbox % Statistical Parametric Mapping (SPM) software https://www.fil.ion.ucl.ac.uk/spm/ % For full functionality, the full suite of CANlab toolboxes is recommended. See here: Installing Tools
% About this dataset % ------------------------------------------------------------------------- % % This dataset includes 33 participants, with brain responses to six levels % of heat (non-painful and painful). % % Aspects of this data appear in these papers: % Wager, T.D., Atlas, L.T., Lindquist, M.A., Roy, M., Choong-Wan, W., Kross, E. (2013). % An fMRI-Based Neurologic Signature of Physical Pain. The New England Journal of Medicine. 368:1388-1397 % (Study 2) % % Woo, C. -W., Roy, M., Buhle, J. T. & Wager, T. D. (2015). Distinct brain systems % mediate the effects of nociceptive input and self-regulation on pain. PLOS Biology. 13(1): % e1002036. doi:10.1371/journal.pbio.1002036 % % Lindquist, Martin A., Anjali Krishnan, Marina López-Solà, Marieke Jepma, Choong-Wan Woo, % Leonie Koban, Mathieu Roy, et al. 2015. ?Group-Regularized Individual Prediction: % Theory and Application to Pain.? NeuroImage. % http://www.sciencedirect.com/science/article/pii/S1053811915009982. %
Load dataset with images and print descriptives
This dataset is shared on figshare.com, under this link: https://figshare.com/s/ca23e5974a310c44ca93
Here is a direct link to the dataset file with the fmri_data object: https://ndownloader.figshare.com/files/12708989
The key variable is image_obj This is an fmri_data object from the CANlab Core Tools repository for neuroimaging data analysis. See https://canlab.github.io/
image_obj.dat contains brain data for each image (average across trials) image_obj.Y contains pain ratings (one average rating per image)
image_obj.additional_info.subject_id contains integers coding for which Load the data file, downloading from figshare if needed
fmri_data_file = which('bmrk3_6levels_pain_dataset.mat'); if isempty(fmri_data_file) % attempt to download disp('Did not find data locally...downloading data file from figshare.com') fmri_data_file = websave('bmrk3_6levels_pain_dataset.mat', 'https://ndownloader.figshare.com/files/12708989'); end load(fmri_data_file); descriptives(image_obj);
Source: BMRK3 dataset from CANlab, PI Tor Wager
Data: .dat contains 6 images per participant, activation estimates during heat on L arm from level 1(44.3 degrees C) to level 6(49.3), in 1 degree increments.
____________________________________________________________________________________________________________________________________________
Wager, T.D., Atlas, L.T., Lindquist, M.A., Roy, M., Choong-Wan, W., Kross, E. (2013). An fMRI-Based Neurologic Signature of Physical Pain.
The New England Journal of Medicine. 368:1388-1397 (Study 2)
Woo, C. -W., Roy, M., Buhle, J. T. & Wager, T. D. (2015). Distinct brain systems mediate the effects of nociceptive input and
self-regulation on pain. PLOS Biology. 13(1): e1002036. doi:10.1371/journal.pbio.1002036
Lindquist, Martin A., Anjali Krishnan, Marina López-Solà, Marieke Jepma, Choong-Wan Woo, Leonie Koban, Mathieu Roy, et al. 2015.
“Group-Regularized Individual Prediction: Theory and Application to Pain.” NeuroImage.
http://www.sciencedirect.com/science/article/pii/S1053811915009982.
____________________________________________________________________________________________________________________________________________
Summary of dataset
______________________________________________________
Images: 198 Nonempty: 198 Complete: 198
Voxels: 223707 Nonempty: 223707 Complete: 223707
Unique data values: 35583281
Min: -11.529 Max: 7.862 Mean: -0.004 Std: 0.227
Percentiles Values
___________ __________
0.1 -1.529
0.5 -0.88055
1 -0.67632
5 -0.31891
25 -0.087519
50 0.00034864
75 0.087461
95 0.29757
99 0.60449
99.5 0.77676
99.9 1.3177
Collect some variables we need
% subject_id is useful for cross-validation % % this used as a custom holdout set in fmri_data.predict() below will % implement leave-one-subject-out cross-validation subject_id = image_obj.additional_info.subject_id; % ratings: reconstruct a subjects x temperatures matrix % so we can plot it % % The command below does this only because we have exactly 6 conditions % nested within 33 subjects and no missing data. ratings = reshape(image_obj.Y, 6, 33)'; % temperatures are 44 - 49 (actually 44.3 - 49.3) in order for each person. temperatures = image_obj.additional_info.temperatures;
Plot the ratings
-------------------------------------------------------------------------
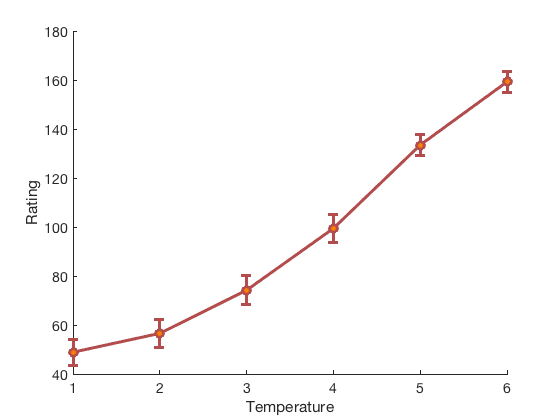
create_figure('ratings'); lineplot_columns(ratings, 'color', [.7 .3 .3], 'markerfacecolor', [1 .5 0]); xlabel('Temperature'); ylabel('Rating');
Prediction
There are many options
% Base model: Linear, whole brain prediction % % Outcome distribution: % Continuous: Regression (LASSO-PCR) Categorical/2 choice: SVM, logistic regression % % Model Options: % - Feature selection (part of the brain) % - Feature integration (new features) % % Testing options: % - Input data (what level) % - Cross-validation % - What testing metric (classification accuracy? RMSE?) and at what level % (single-trial, condition averages, one-map-per-subject) % % Inference options: % - Component maps, voxel-wise maps % - Thresholding (Bootstrapping, Permutation) % - Global null hypothesis testing / sanity checks (Permutation)
Run the Base model
% relevant functions: % predict method (fmri_data) % predict_test_suite method (fmri_data) % Define custom holdout set. If we use subject_id, which is a vector of % integers with a unique integer per subject, then we are doing % leave-one-subject-out cross-validation. % let's build five-fold cross-validation set that leaves out ALL the images % from a subject together. That way, we are always predicting out-of-sample % (new individuals). If not, dependence across images from the same % subjects may invalidate the estimate of predictive accuracy. holdout_set = zeros(size(subject_id)); % holdout set membership for each image n_subjects = length(unique(subject_id)); C = cvpartition(n_subjects, 'KFold', 5); for i = 1:5 teidx = test(C, i); % which subjects to leave out imgidx = ismember(subject_id, find(teidx)); % all images for these subjects holdout_set(imgidx) = i; end algoname = 'cv_lassopcr'; % cross-validated penalized regression. Predict pain ratings [cverr, stats, optout] = predict(image_obj, 'algorithm_name', algoname, 'nfolds', holdout_set);
Cross-validated prediction with algorithm cv_lassopcr, 5 folds Completed fit for all data in: 0 hours 0 min 5 secs Fold 1/5 done in: 0 hours 0 min 4 sec Fold 2/5 done in: 0 hours 0 min 4 sec Fold 3/5 done in: 0 hours 0 min 3 sec Fold 4/5 done in: 0 hours 0 min 3 sec Fold 5/5 done in: 0 hours 0 min 4 sec Total Elapsed Time = 0 hours 0 min 22 sec
Plot the results
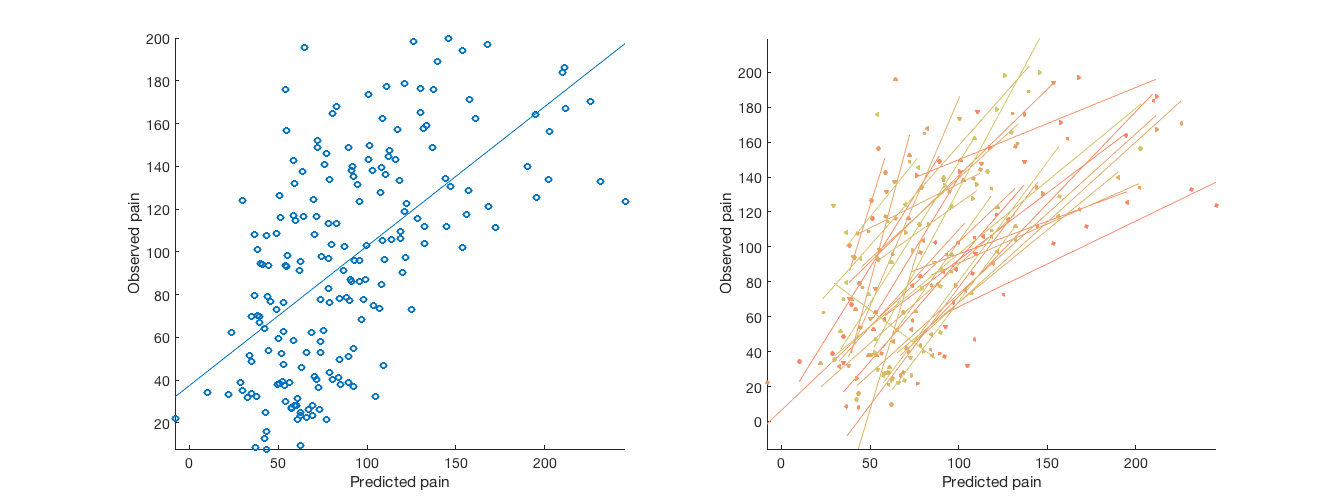
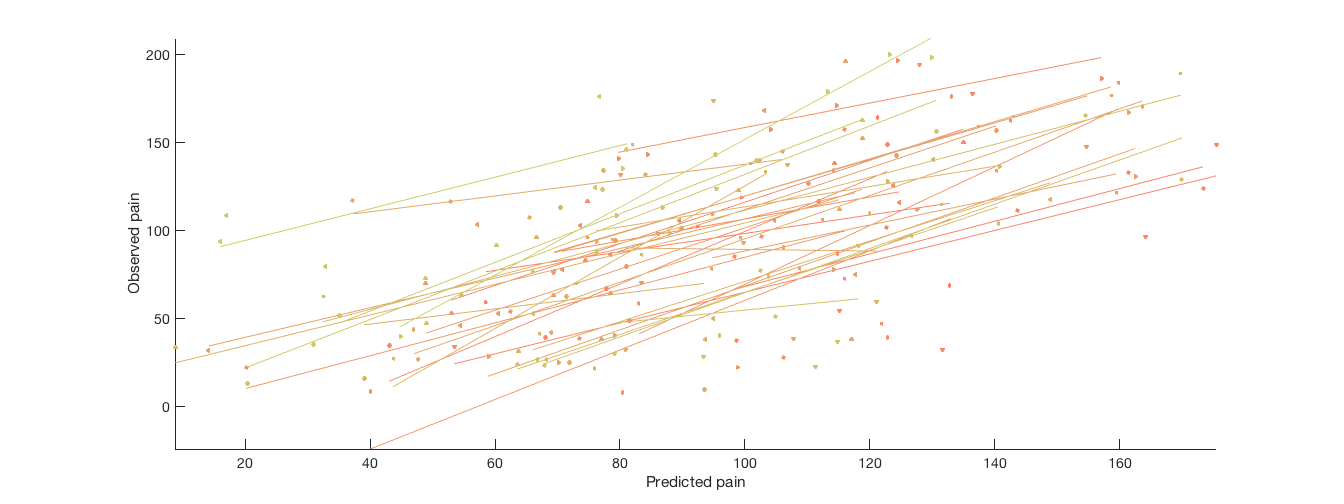
% Plot cross-validated predicted outcomes vs. actual % Critical fields in stats output structure: % stats.Y = actual outcomes % stats.yfit = cross-val predicted outcomes % pred_outcome_r: Correlation between .yfit and .Y % weight_obj: Weight map used for prediction (across all subjects). % Continuous outcomes: create_figure('scatterplots', 1, 2); plot(stats.yfit, stats.Y, 'o') axis tight refline xlabel('Predicted pain'); ylabel('Observed pain'); % Consider that each subject has their own line: for i = 1:n_subjects YY{i} = stats.Y(subject_id == i); Yfit{i} = stats.yfit(subject_id == i); end subplot(1, 2, 2); line_plot_multisubject(Yfit, YY, 'nofigure'); xlabel('Predicted pain'); ylabel('Observed pain'); axis tight % Yfit is a cell array, one cell per subject, with fitted values for that subject
average within-person r = 0.60, t( 32) = 7.93, p = 0.000000, num. missing: 0
Summarize within-person classification accuracy
Turn into classification:
% If the stim intensity increases by 1 degree, how often do we correctly % predict an increase? diffs = cellfun(@diff, Yfit, 'UniformOutput', false); % successive increases in predicted values with increases in stim intensity % Subjects (rows) x comparisons (cols), each comparison 1 degree apart diffs = cat(2, diffs{:})'; acc_per_subject = sum(diffs > 0, 2) ./ size(diffs, 2); acc_per_comparison = (sum(diffs > 0) ./ size(diffs, 1))'; cohens_d_per_comparison = (mean(diffs) ./ std(diffs))'; fprintf('Mean acc is : %3.2f%% across all successive 1 degree increments\n', 100*mean(acc_per_subject)); % Create and display a table: comparison = {'45-44' '46-45' '47-46' '48-47' '49-48'}'; results_table = table(comparison, acc_per_comparison, cohens_d_per_comparison); disp(results_table) % 45-44 degrees, 46-45, 47-46, etc.
Mean acc is : 73.94% across all successive 1 degree increments
comparison acc_per_comparison cohens_d_per_comparison
__________ __________________ _______________________
'45-44' 0.48485 -0.0587
'46-45' 0.81818 0.76512
'47-46' 0.84848 0.93114
'48-47' 0.9697 1.432
'49-48' 0.57576 0.37981
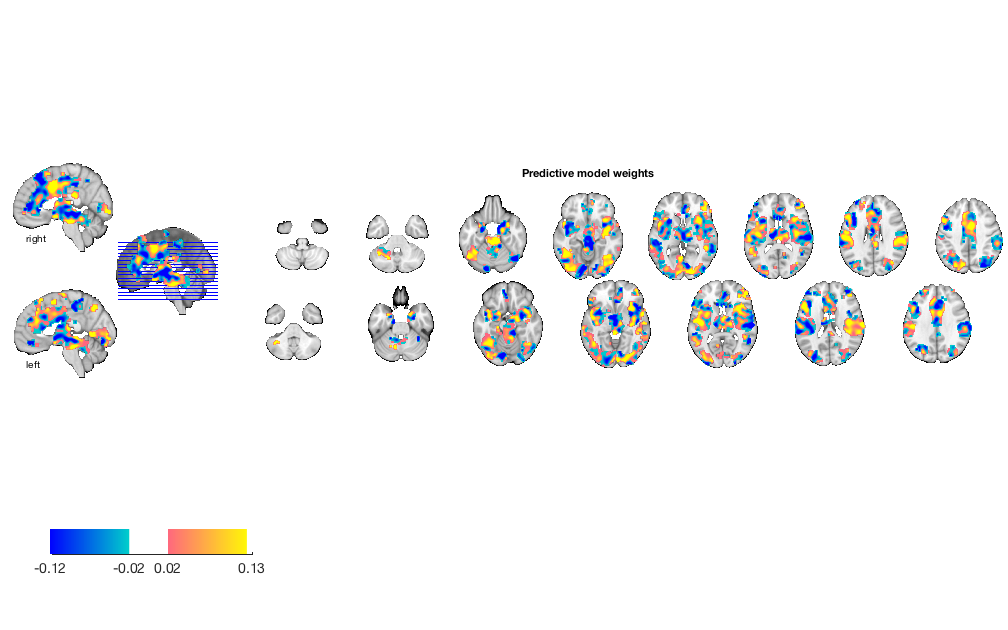
Visualize weight map
w = stats.weight_obj; % This is an image object that we can view, etc. orthviews(w) create_figure('montage'); o2 = montage(w); o2 = title_montage(o2, 5, 'Predictive model weights');
SPM12: spm_check_registration (v6245) 17:48:58 - 03/08/2018
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
Grouping contiguous voxels: 3 regions
ans =
1×1 cell array
{1×3 region}
Setting up fmridisplay objects
This takes a lot of memory, and can hang if you have too little.
Grouping contiguous voxels: 3 regions
sagittal montage: 4350 voxels displayed, 219357 not displayed on these slices
sagittal montage: 4292 voxels displayed, 219415 not displayed on these slices
sagittal montage: 3990 voxels displayed, 219717 not displayed on these slices
axial montage: 32223 voxels displayed, 191484 not displayed on these slices
axial montage: 34538 voxels displayed, 189169 not displayed on these slices
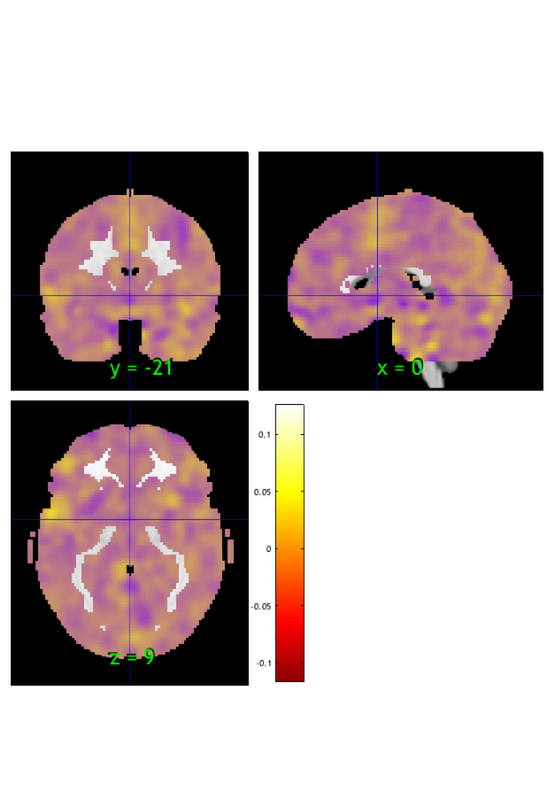
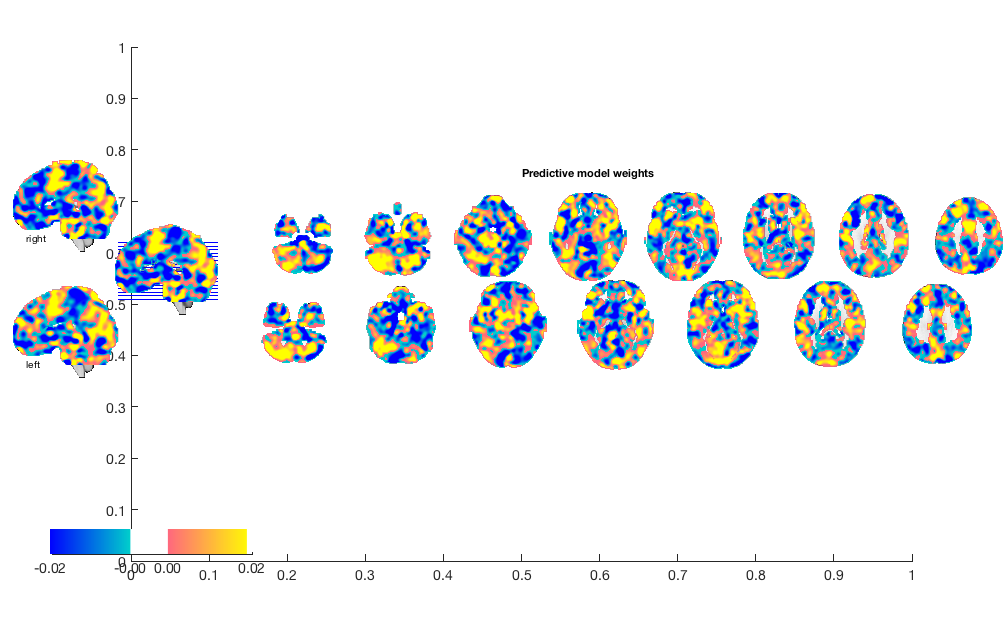
Bootstrap weights: Get most reliable weights and p-values for voxels
% Re-run prediction, adding a 'bootstrap' flag. % For a final analysis, at least 5K bootstrap samples is a good idea % (This is not run in the example; see help fmri_data.predict for how to % run bootstrapping)
Try normalizing/scaling data
% z-score every image, removing the mean and dividing by the std. % Remove some scaling noise, but also some signal... image_obj = rescale(image_obj, 'zscoreimages'); % re-run prediction and plot [cverr, stats, optout] = predict(image_obj, 'algorithm_name', algoname, 'nfolds', holdout_set); create_figure('scatterplots'); for i = 1:n_subjects YY{i} = stats.Y(subject_id == i); Yfit{i} = stats.yfit(subject_id == i); end line_plot_multisubject(Yfit, YY, 'nofigure'); xlabel('Predicted pain'); ylabel('Observed pain'); axis tight fprintf('Overall prediction-outcome correlation is %3.2f\n', stats.pred_outcome_r) figure; o2 = montage(stats.weight_obj); o2 = title_montage(o2, 5, 'Predictive model weights');
Cross-validated prediction with algorithm cv_lassopcr, 5 folds Completed fit for all data in: 0 hours 0 min 5 secs Fold 1/5 done in: 0 hours 0 min 4 sec Fold 2/5 done in: 0 hours 0 min 3 sec Fold 3/5 done in: 0 hours 0 min 3 sec Fold 4/5 done in: 0 hours 0 min 3 sec Fold 5/5 done in: 0 hours 0 min 4 sec Total Elapsed Time = 0 hours 0 min 22 sec average within-person r = 0.59, t( 32) = 13.18, p = 0.000000, num. missing: 0 Overall prediction-outcome correlation is 0.59 Setting up fmridisplay objects This takes a lot of memory, and can hang if you have too little. Grouping contiguous voxels: 3 regions sagittal montage: 4350 voxels displayed, 219357 not displayed on these slices sagittal montage: 4292 voxels displayed, 219415 not displayed on these slices sagittal montage: 3990 voxels displayed, 219717 not displayed on these slices axial montage: 32223 voxels displayed, 191484 not displayed on these slices axial montage: 34538 voxels displayed, 189169 not displayed on these slices
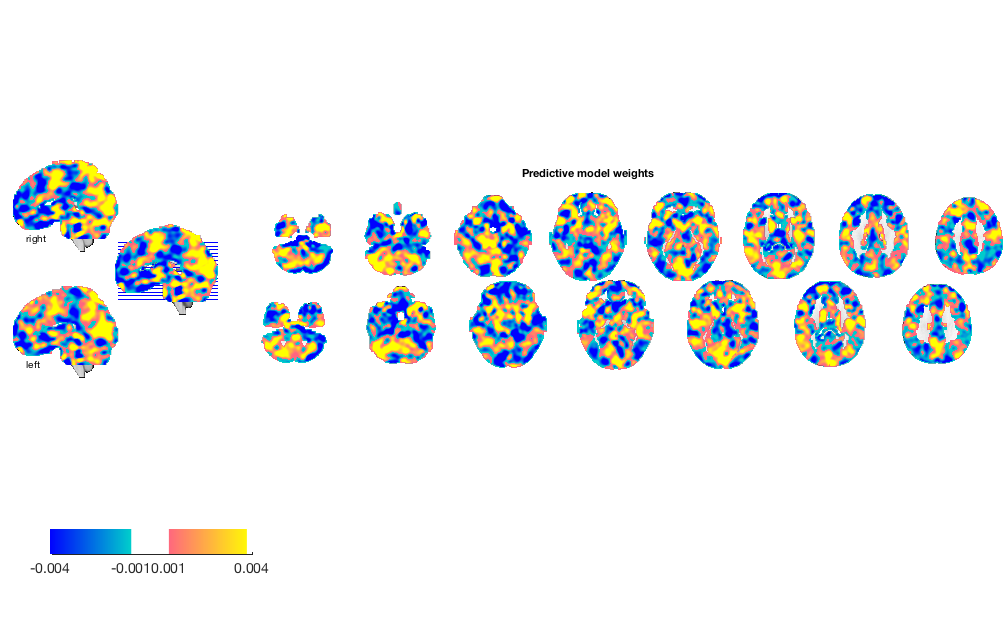
Try selecting an a priori 'network of interest'
% re-load load('bmrk3_6levels_pain_dataset.mat', 'image_obj') % load an a priori pain-related mask and apply it mask = fmri_data(which('pain_2s_z_val_FDR_05.img.gz')); % in Neuroimaging_Pattern_Masks repository image_obj = apply_mask(image_obj, mask); % show mean data with mask m = mean(image_obj); figure; o2 = montage(m); o2 = title_montage(o2, 5, 'Mean image data, masked');
Using default mask: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/brainmask.nii loading mask. mapping volumes. checking that dimensions and voxel sizes of volumes are the same. Pre-allocating data array. Needed: 1409312 bytes Loading image number: 1 Elapsed time is 0.019081 seconds. Image names entered, but fullpath attribute is empty. Getting path info. Setting up fmridisplay objects This takes a lot of memory, and can hang if you have too little. Grouping contiguous voxels: 27 regions sagittal montage: 1088 voxels displayed, 47895 not displayed on these slices sagittal montage: 1171 voxels displayed, 47812 not displayed on these slices sagittal montage: 980 voxels displayed, 48003 not displayed on these slices axial montage: 7496 voxels displayed, 41487 not displayed on these slices axial montage: 7950 voxels displayed, 41033 not displayed on these slices
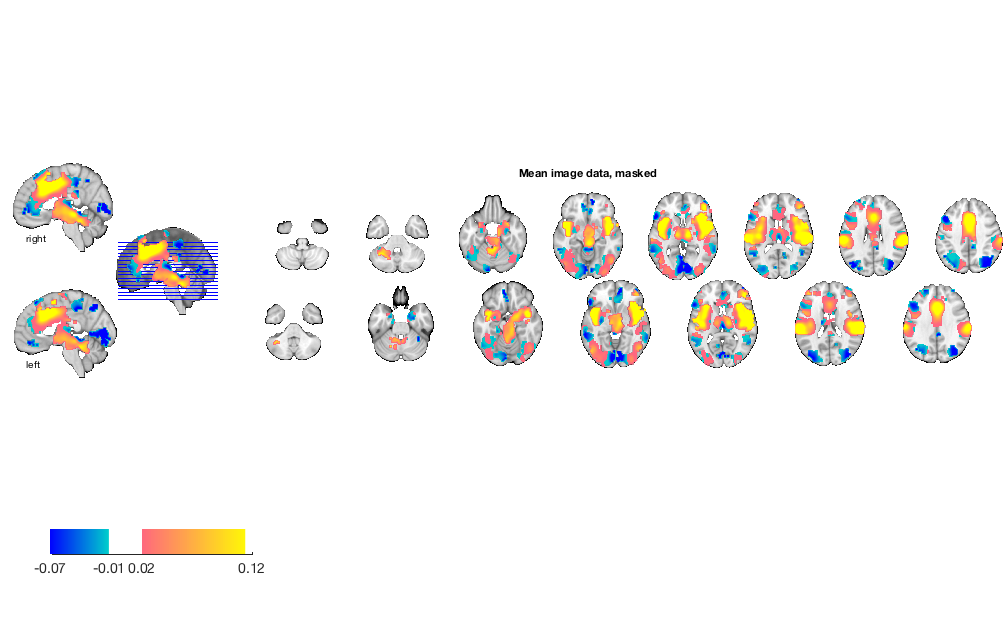
re-run prediction and plot
[cverr, stats, optout] = predict(image_obj, 'algorithm_name', algoname, 'nfolds', holdout_set); create_figure('scatterplots'); for i = 1:n YY{i} = stats.Y(subject_id == i); Yfit{i} = stats.yfit(subject_id == i); end line_plot_multisubject(Yfit, YY, 'nofigure'); xlabel('Predicted pain'); ylabel('Observed pain'); axis tight fprintf('Overall prediction-outcome correlation is %3.2f\n', stats.pred_outcome_r) figure; o2 = montage(stats.weight_obj); o2 = title_montage(o2, 5, 'Predictive model weights');
Cross-validated prediction with algorithm cv_lassopcr, 5 folds Completed fit for all data in: 0 hours 0 min 1 secs Fold 1/5 done in: 0 hours 0 min 1 sec Fold 2/5 done in: 0 hours 0 min 1 sec Fold 3/5 done in: 0 hours 0 min 1 sec Fold 4/5 done in: 0 hours 0 min 1 sec Fold 5/5 done in: 0 hours 0 min 1 sec Total Elapsed Time = 0 hours 0 min 4 sec average within-person r = 0.60, t( 32) = 6.59, p = 0.000000, num. missing: 0 Overall prediction-outcome correlation is 0.60 Setting up fmridisplay objects This takes a lot of memory, and can hang if you have too little. Grouping contiguous voxels: 27 regions sagittal montage: 1088 voxels displayed, 47895 not displayed on these slices sagittal montage: 1171 voxels displayed, 47812 not displayed on these slices sagittal montage: 980 voxels displayed, 48003 not displayed on these slices axial montage: 7496 voxels displayed, 41487 not displayed on these slices axial montage: 7950 voxels displayed, 41033 not displayed on these slices
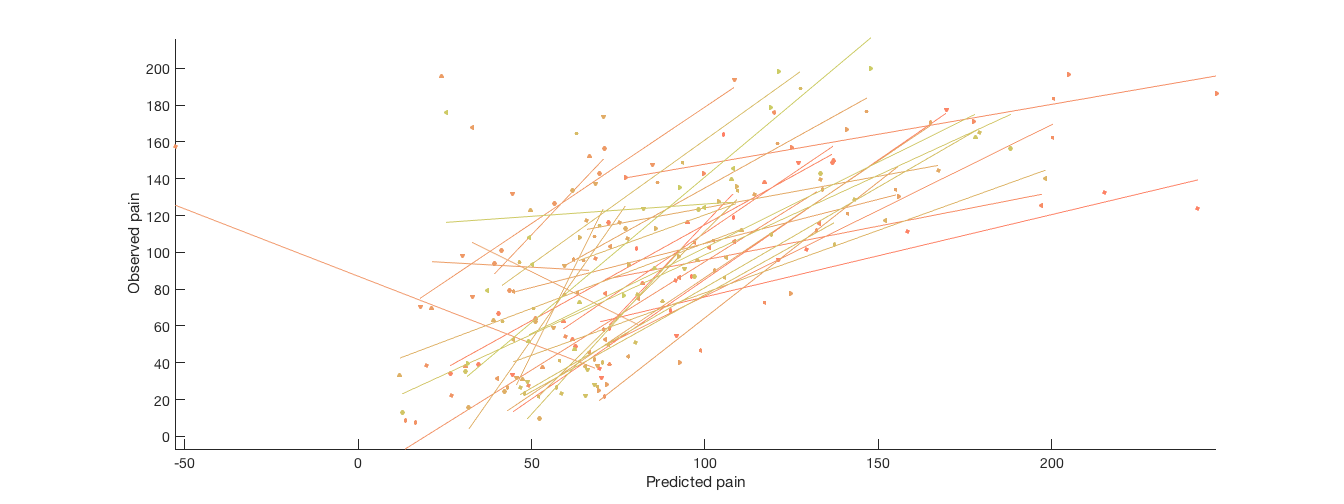
Other ideas
There are many ways to build from this basic analysis.
% Data % ------------ % Examine/clean outliers % Transformations of data: Scaling/centering to remove global mean, % ventricle mean % Identify unaccounted sources of variation/subgroups (experimenter sex, % etc.) % Features % ------------ % A priori regions / meta-analysis % Feature selection in cross-val loop: Univariate, recursive feature elim