Contents
Mediation analysis basic walkthrough
This script is explained in more detail in the powerpoint titled Mediation_sample_data_walkthrough
% mediation_example_script1 and 2 do the same analysis. % ...script1 is very terse, and includes the essential commands only. % ...script2 is longer and includes more checking that files are available, etc.
Step 1: Make a new analysis directory to save results, and go there
Make a new analysis directory to save results, and go there
andir = 'Test_mediation';
mkdir(andir)
cd(andir)
Step 2: Load image data and behavioral variables
dinf = what('Wager_et_al_2008_Neuron_EmotionReg'); %imgs = filenames(fullfile(dinf.path,'con_*img'), 'char', 'absolute'); imgs = fullfile(dinf.path, 'Wager_2008_emo_reg_vs_look_neg_contrast_images.nii.gz'); behav_dat = importdata(fullfile(dinf.path,'Wager_2008_emotionreg_behavioral_data.txt'))
behav_dat =
struct with fields:
data: [30×2 double]
textdata: {'X_RVLPFC' 'Y_Reappraisal_Success'}
colheaders: {'X_RVLPFC' 'Y_Reappraisal_Success'}
Step 3: Load and display mask
The mask determines which voxels are analyzed. The standard mask is in the CanlabCore Tools repository, so you need the folder containing it (and other CanlabCore folders) on your path.
mask = which('gray_matter_mask.img') canlab_results_fmridisplay(mask, 'compact2');
mask =
'/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/gray_matter_mask.img'
Using default mask: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/brainmask.nii
loading mask. mapping volumes.
checking that dimensions and voxel sizes of volumes are the same.
Pre-allocating data array. Needed: 1409312 bytes
Loading image number: 1
Elapsed time is 0.019277 seconds.
Image names entered, but fullpath attribute is empty. Getting path info.
Grouping contiguous voxels: 46 regions
Setting up fmridisplay objects
This takes a lot of memory, and can hang if you have too little.
axial montage: 42172 voxels displayed, 169167 not displayed on these slices
sagittal montage: 8208 voxels displayed, 203131 not displayed on these slices
axial montage: 42172 voxels displayed, 169167 not displayed on these slices
sagittal montage: 8208 voxels displayed, 203131 not displayed on these slices

Step 4 : Run mediation
Run mediation without bootstrapping (fast) Test that things are working
x=behav_dat.data(:,1);
y=behav_dat.data(:,2);
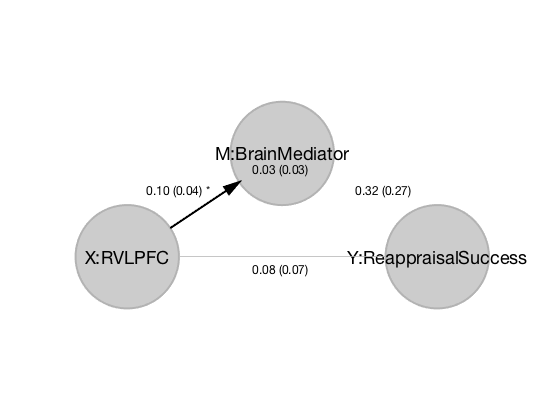
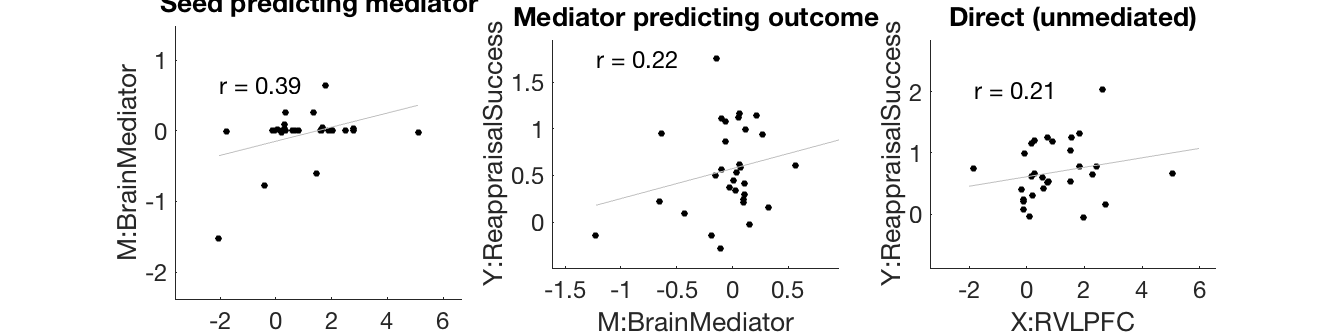
names = {'X:RVLPFC' 'Y:Reappraisal_Success' 'M:BrainMediator'};
% This is what you would run:
% results = mediation_brain(x,y,imgs,'names',names,'mask', mask);
% We run this instead to suppress output for report publishing
str = 'results = mediation_brain(x,y,imgs,''names'',names,''mask'', mask);';
disp(['Running with output suppressed (for report-generation): ' str]);
evalc(str);
% "Legacy" version: reslice mask to same space first:
% scn_map_image(mask,deblank(imgs(1,:)), 'write', 'resliced_mask.img');
% results = mediation_brain(x,y,imgs,'names',names,'mask', 'resliced_mask.img');
% OR
% ---------------------------------
% Uncomment the lines below to run with bootstrapping
% Make yourself a cup of tea while the results are compiled as this is going to take a while
% pre-compiled results are also available in
% 'mediation_Example_Data_Wager2008_Msearch_R_XisRIFGstim_norobust'
% results = mediation_brain(x,y,imgs,'names',names,'mask', mask,'boot','pvals',5, 'bootsamples', 10000);
Running with output suppressed (for report-generation): results = mediation_brain(x,y,imgs,'names',names,'mask', mask);
Step 5: Get and save results figures, tables, and report
First, change to the mediation analysis directory, if you're not there already. Then, run one of several batch results functions.
The most complete way to create and publish a time- and date-stamped HTML report with figures and tables is to run "publish_mediation_report.m" (This is commented out because this script is published, and we can't have nested publish commands)
publish_mediation_report;
--------------------------------------------------------------------- Another option is to use this batch script to create figures and tables, and save .mat files with results:
mediation_brain_results_all_script; % This function runs a series of calls to mediation_brain_results.m, which % is also a stand-alone function. Type "help mediation_brain_results" for more options.
---------------------------------------------- Analysis Mask ---------------------------------------------- Using default anatomical underlay image. montage_clusters: no activation blobs; displaying underlay image. 0 Clusters found. No overlap plotting.

----------------------------------------------
Overlap between Paths a and b
----------------------------------------------
-------------------------------------------------------------------
-------------------------------------------------------------------
MEDIATION: OVERLAP BETWEEN PATH A AND B MAPS
-------------------------------------------------------------------
-------------------------------------------------------------------
Setting up fmridisplay objects
This takes a lot of memory, and can hang if you have too little.
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
axial montage: 3 voxels displayed, 18 not displayed on these slices
axial montage: 5 voxels displayed, 16 not displayed on these slices
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
sagittal montage: 2 voxels displayed, 19 not displayed on these slices
axial montage: 3 voxels displayed, 18 not displayed on these slices
axial montage: 5 voxels displayed, 16 not displayed on these slices
Saving clusters with extracted data in:
intersect_a_and_b_clusters_005_k3
----------------------------------------------
MEDIATION: A, B, A*B CONJUNCTION
----------------------------------------------
-------------------------------------------------------------------
-------------------------------------------------------------------
MEDIATION: A, B, A*B CONJUNCTION
-------------------------------------------------------------------
-------------------------------------------------------------------
Will print tables.
Will show slices.
Creating image XMY_conjunction.img with max p-values for relevant paths
SPM12: spm_check_registration (v6245) 13:49:38 - 10/01/2019
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
(<a href="matlab:spm_check_registration('/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">all</a>) <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
<a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
<a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
Saving log file: XMY_conjunction_005_01_05_k3_1_1_prune_log.txt
Other key results in: XMY_conjunction_005_01_05_k3_1_1_prune_results.txt
Will also save figures of slice montages if 'slices' requested.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
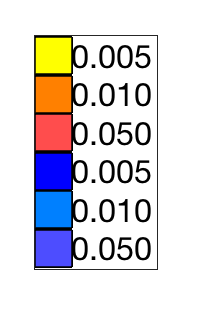
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
----------------------
No significant results
----------------------
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
----------------------
No significant results
----------------------
Extracting image data for significant clusters. No clusters to extract.
Extracting image data for significant clusters. No clusters to extract.
Summary of output images:
/Users/torwager/Downloads/Test_mediation/Test_mediation/Test_mediation/X-M_pvals.img
/Users/torwager/Downloads/Test_mediation/Test_mediation/Test_mediation/M-Y_pvals.img
/Users/torwager/Downloads/Test_mediation/Test_mediation/Test_mediation/X-M-Y_pvals.img
/Users/torwager/Downloads/Test_mediation/Test_mediation/Test_mediation/XMY_conjunction.img
Results clusters clpos and clneg are returned for the LAST image in this set.
Printing Tables.
Saving clusters in cl_XMY_conjunction_005_01_05_k3_1_1_prune
No clusters to show slices for.

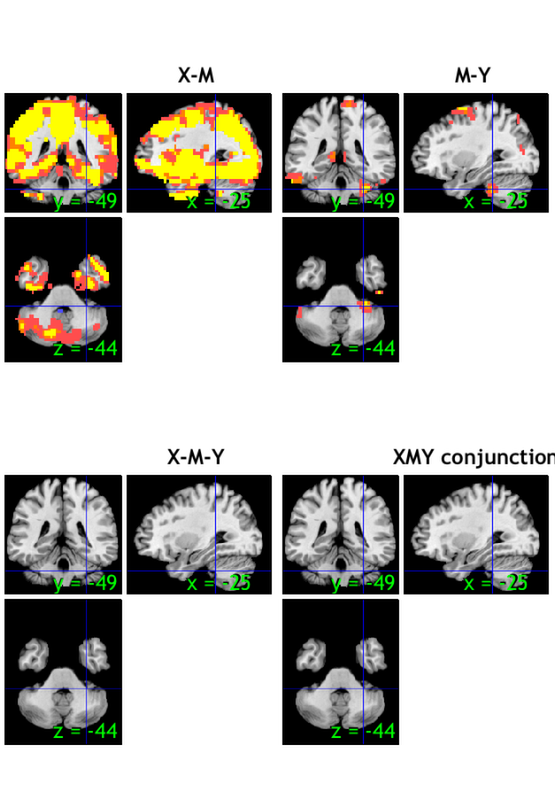
-------------------------------------------------------------------
-------------------------------------------------------------------
MEDIATION: PATH A
-------------------------------------------------------------------
-------------------------------------------------------------------
Will print tables.
Will show slices.
Saving log file: X-M_pvals_005_01_05_k3_1_1_prune_log.txt
Other key results in: X-M_pvals_005_01_05_k3_1_1_prune_results.txt
Will also save figures of slice montages if 'slices' requested.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
SPM12: spm_check_registration (v6245) 13:49:55 - 10/01/2019
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
Extracting image data for significant clusters. Images are not valid files.
Extracting image data for significant clusters. Images are not valid files.
Summary of output images:
X-M_pvals.img
Results clusters clpos and clneg are returned for the LAST image in this set.
Printing Tables.
NOTE: FULL MEDIATION TABLES DO NOT WORK YET FOR SINGLE-LEVEL RESULTS
Printing abbreviated tables.
Positive effects
Z field contains: Mediation a effect (shown in maxstat)
Name index x y z corr voxels volume_mm3 maxstat
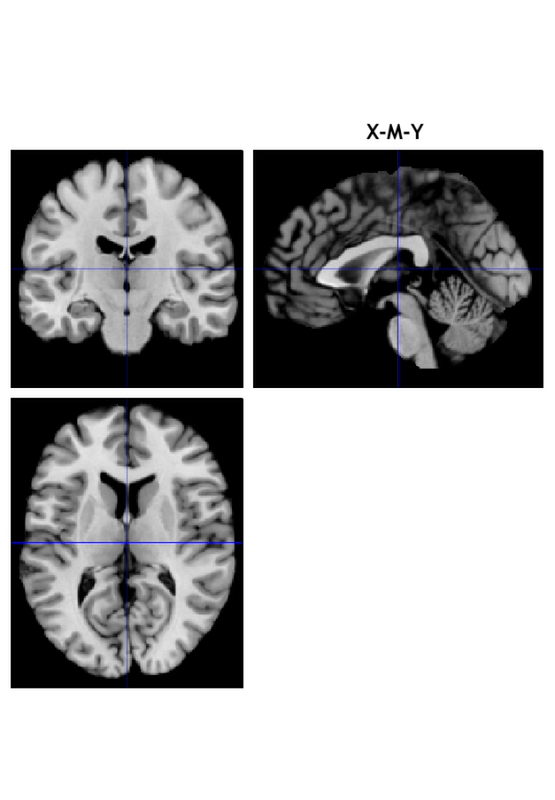
R1 1 -3 -17 18 NaN 24151 -1284201 32.91
Negative effects
Z field contains: Mediation a effect (shown in maxstat)
Name index x y z corr voxels volume_mm3 maxstat
R1 1 0 -48 -45 NaN 33 -1755 6.12
Saving clusters in cl_X-M_pvals_005_01_05_k3_1_1_prune
Saving image of slices: orth_X-M_pvals_005_01_05_k3_1_1_prune_coronal.png
Saving image of slices: orth_X-M_pvals_005_01_05_k3_1_1_prune_sagittal.png
Saving image of slices: orth_X-M_pvals_005_01_05_k3_1_1_prune_axial.png

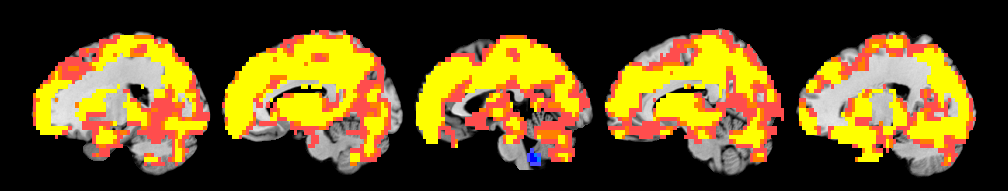
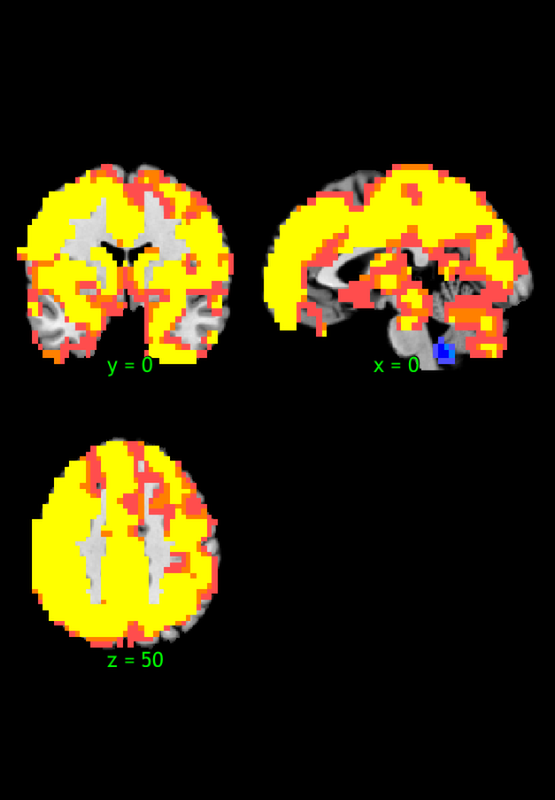

-------------------------------------------------------------------
-------------------------------------------------------------------
MEDIATION: PATH B
-------------------------------------------------------------------
-------------------------------------------------------------------
Will print tables.
Will show slices.
Saving log file: M-Y_pvals_005_01_05_k3_1_1_prune_log.txt
Other key results in: M-Y_pvals_005_01_05_k3_1_1_prune_results.txt
Will also save figures of slice montages if 'slices' requested.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
SPM12: spm_check_registration (v6245) 13:50:18 - 10/01/2019
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
Extracting image data for significant clusters. Images are not valid files.
Extracting image data for significant clusters. No clusters to extract.
Summary of output images:
M-Y_pvals.img
Results clusters clpos and clneg are returned for the LAST image in this set.
Printing Tables.
NOTE: FULL MEDIATION TABLES DO NOT WORK YET FOR SINGLE-LEVEL RESULTS
Printing abbreviated tables.
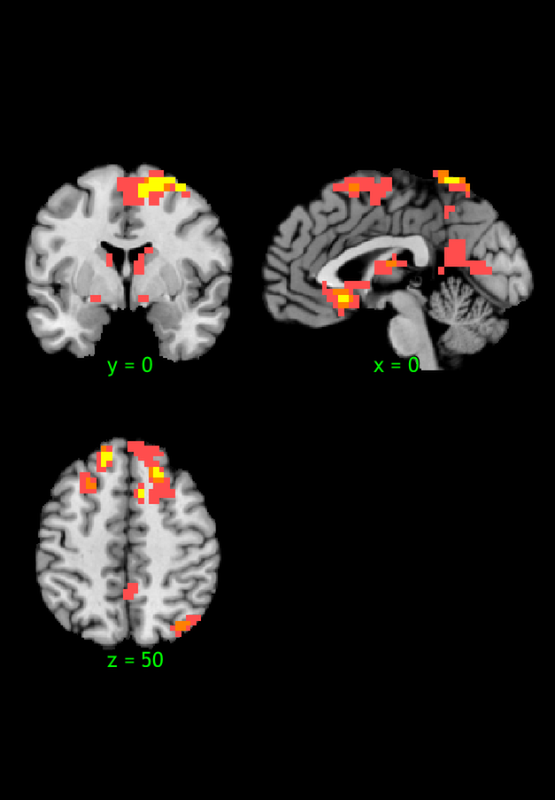
Positive effects
Z field contains: Mediation b effect (shown in maxstat)
Name index x y z corr voxels volume_mm3 maxstat
R1 1 31 -38 -36 NaN 56 -2978 6.23
R2 2 -48 -34 -22 NaN 159 -8455 7.88
R3 3 52 -28 -32 NaN 48 -2552 7.36
R4 4 0 3 0 NaN 552 -29352 7.68
R5 5 41 -72 32 NaN 168 -8933 6.01
R6 6 7 17 54 NaN 640 -34031 8.55
R7 7 3 -48 68 NaN 116 -6168 8.52
Negative effects
No results to print.
Saving clusters in cl_M-Y_pvals_005_01_05_k3_1_1_prune
Saving image of slices: orth_M-Y_pvals_005_01_05_k3_1_1_prune_coronal.png
Saving image of slices: orth_M-Y_pvals_005_01_05_k3_1_1_prune_sagittal.png
Saving image of slices: orth_M-Y_pvals_005_01_05_k3_1_1_prune_axial.png

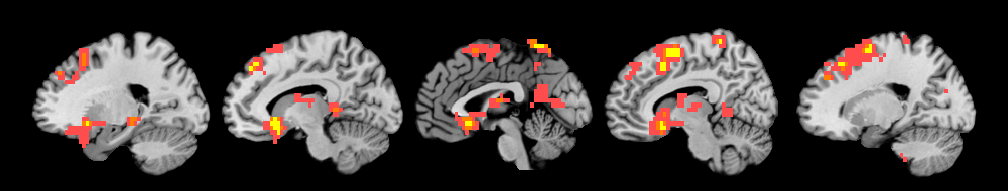

-------------------------------------------------------------------
-------------------------------------------------------------------
MEDIATION: PATH A*B
-------------------------------------------------------------------
-------------------------------------------------------------------
Will print tables.
Will show slices.
Saving log file: X-M-Y_pvals_005_01_05_k3_1_1_prune_log.txt
Other key results in: X-M-Y_pvals_005_01_05_k3_1_1_prune_results.txt
Will also save figures of slice montages if 'slices' requested.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: pos
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
Cannot find SPM graphics window. It should have a tag of 'Graphics'.
iimg_multi_threshold viewer
=====================================================
Showing positive or negative: neg
Overlay is: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img
Mask is entered in iimg_multi_threshold as a vector of numerical values
Entered p-value image? : Yes
Height thresholds:0.0050 0.0100 0.0500
Extent thresholds: 3 1 1
Show only contiguous with seed regions: Yes
Warning: p-values in cl.Z will not give valid spm_max subclusters.
log(1/p) saved in output cl.Z field.
Cannot find SPM graphics window. It should have a tag of 'Graphics'.
----------------------
No significant results
----------------------
SPM12: spm_check_registration (v6245) 13:50:47 - 10/01/2019
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/SPM8_colin27T1_seg.img,1</a>
Extracting image data for significant clusters. No clusters to extract.
Extracting image data for significant clusters. No clusters to extract.
Summary of output images:
X-M-Y_pvals.img
Results clusters clpos and clneg are returned for the LAST image in this set.
Printing Tables.
Saving clusters in cl_X-M-Y_pvals_005_01_05_k3_1_1_prune
No clusters to show slices for.