Load and explore a sample dataset
This example shows how to load a pre-set example fMRI dataset into an fmri_data object.
Contents
- General instructions
- The fmri_data object
- Section 1: The quick and easy way to load a pre-specified dataset
- Section 2: Manual load. Use filenames to find file names
- Section 3: Plot the data we just loaded
- Section 4: Get help on an object
- Section 5: Explore methods
- Section 6: Explore on your own
- Explore More: CANlab Toolboxes
General instructions
Before you start, the CANlab_Core_Tools must be added to your path with subfolders. Otherwise, you will get errors.
The script canlab_toolbox_setup can help download and install the toolboxes you need.
Sample datasets are in the "Sample_datasets" folder in CANlab_Core_Tools.
This example will use emotion regulation data in the folder: "Wager_et_al_2008_Neuron_EmotionReg" The dataset is a series of contrast images from N = 30 participants. Each image is a contrast image for [reappraise neg vs. look neg]
These data were published in: Wager, T. D., Davidson, M. L., Hughes, B. L., Lindquist, M. A., Ochsner, K. N.. (2008). Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron, 59, 1037-50.
Here are a couple of helpful functions we will use for display: (you can ignore these.)
dashes = '----------------------------------------------'; printhdr = @(str) fprintf('%s\n%s\n%s\n', dashes, str, dashes);
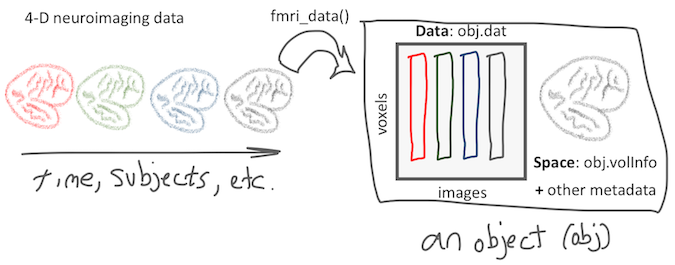
The fmri_data object
The fmri_data object class is one of the most important, basic types of objects in the CANlab object-oriented toolbox. It stores image data in a matrix form, which is more space efficient and friendly for analysis with various software packages/algorithms.
For philosophy, see: https://canlab.github.io/
For more info on the object-oriented approach, see: https://canlab.github.io/objectoriented/
When you call the class constructor fmri_data(), this is what it does:
Section 1: The quick and easy way to load a pre-specified dataset
The function load_image_set has a number of pre-defined image sets that you can load with one simple command. This is the easiest way to load sample data. The images must be on your Matlab path for this to work.
[data_obj, subject_names, image_names] = load_image_set('emotionreg'); % data_obj is an fmri_data object containing all 30 images. % subject_names is a list of short names for each image. % image_names is a list of the full image names with their path.
Loaded images: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/Sample_datasets/Wager_et_al_2008_Neuron_EmotionReg/Wager_2008_emo_reg_vs_look_neg_contrast_images.nii
Section 2: Manual load. Use filenames to find file names
First, we need to list the file names in a string matrix. Then, we can load them into an fmri_data object We will use the filenames function to get the names, and the fmri_data object constructor function to load the data.
% First, check whether images are on your path: % We will search for one image and save the path name. printhdr('Check that we can find data images:') myfile = which('con_00810001.img'); mydir = fileparts(myfile); if isempty(mydir), disp('Uh-oh! I can''t find the data.'), else disp('Data found.'), end % Now we can list all the file names. printhdr('Find files and get their names:') image_names = filenames(fullfile(mydir, 'con_008100*img'), 'absolute'); disp('Done.'); % Now load them into an fmri_data object. printhdr('Loading the image data into the object:') data_obj = fmri_data(image_names); % This is the gateway to doing many other things, which are explained in % other help files. But just to get us started, let's run through a few % basic things we can do. We'll mainly just look at some standard plots of % the data.
---------------------------------------------- Check that we can find data images: ---------------------------------------------- Data found. ---------------------------------------------- Find files and get their names: ---------------------------------------------- Done. ---------------------------------------------- Loading the image data into the object: ---------------------------------------------- Using default mask: /Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/brainmask.nii loading mask. mapping volumes. checking that dimensions and voxel sizes of volumes are the same. Pre-allocating data array. Needed: 5975040 bytes Loading image number: 30 Elapsed time is 0.180418 seconds. fmri_data.create: Converting 1203 NaNs to 0s.Image names entered, but fullpath attribute is empty. Getting path info.
Section 3: Plot the data we just loaded
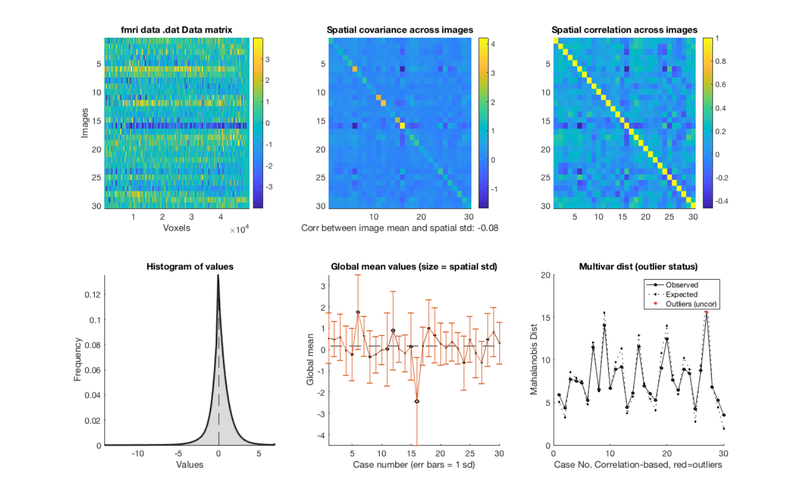
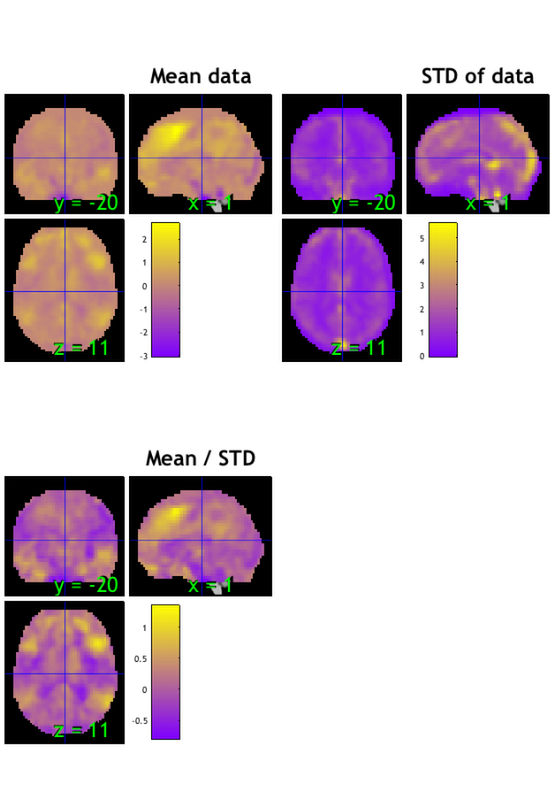
Operations that we can perform on fmri_data objects are called methods. You can see a list of methods by typing methods(data_obj). Here, we'll call the plot method to visualize the data.
plot(data_obj) snapnow
Retained 8 components for mahalanobis distance
Expected 50% of points within 50% normal ellipsoid, found 46.67%
Expected 1.50 outside 95% ellipsoid, found 1
Potential outliers based on mahalanobis distance:
Bonferroni corrected: 0 images Cases
Uncorrected: 1 images Cases 27
Outliers:
Outliers after p-value correction:
Image numbers:
Image numbers, uncorrected: 27
Warning: Ignoring extra legend entries.
SPM12: spm_check_registration (v6245) 17:02:02 - 09/01/2020
========================================================================
Display <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
(<a href="matlab:spm_check_registration('/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">all</a>) <a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
<a href="matlab:spm_image('display','/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1');">/Users/torwager/Documents/GitHub/CanlabCore/CanlabCore/canlab_canonical_brains/Canonical_brains_surfaces/keuken_2014_enhanced_for_underlay.img,1</a>
ans =
30×1 logical array
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
1
0
0
0
Section 4: Get help on an object
Objects have associated help files. Type:
help fmri_data % ...to get help for the fmri_data() class constructor and object. % % Some objects also have doc files that contain additional information. % Try: doc fmri_data
fmri_data: Data class for storing and analyzing neuroimaging datasets and associated information
'fmri_data' is a data class containing information about generic fmri
datasets stored in a structure-like object. Using this has the
advantages that the properties and methods are standardized and controlled.
It also keeps track of the history of what was done to the dataset.
-------------------------------------------------------------------------
Features and philosophy:
-------------------------------------------------------------------------
- Store image data in a flat (2-d), space-efficient voxels x images matrix
- Save space: Tools to remove out-of-image voxels and empty (zero/NaN) voxels, store data in single-precision format
- Analysis-friendly: 2-d matrices can be read and analyzed in multiple packages/algorithms
- Meta-data included to convert back to 3-d image volume space,
with easy tools (methods) to reconstruct and visualize images
Built-in resampling makes it easy to compare/combine datasets with different voxel sizes and image bounding boxes
Reduces overhead for statisticians/data scientists unfamiliar with neuroimaging to apply their algorithms.
- Multiple images can be stored in a single object
- Methods have short, intuitive names, and perform high-level functions specialized for neuroimaging:
- Visualization (plot, orthviews, surface, montage, histogram, isosurface methods)
- Image manipulation (apply_mask, get_wh_image, resample_space, compare_space, flip, threshold methods)
- Data extraction (apply_atlas, apply_parcellation, extract_gray_white_csf, extract_roi_averages)
- Analysis (ica, mahal, image_math, and many more in the fmri_data subclass)
- Provenance: Ability to track and update history of changes to objects
fmri_data is a subclass of the image_vector object and inherits its
properties and methods.
-------------------------------------------------------------------------
To construct/create a new instance of an object:
-------------------------------------------------------------------------
Basic usage:
obj = fmri_data(image_names, [maskinput], [other optional inputs])
maskinput : [optional] name of mask image to use. Default: 'brainmask.nii', a
brain mask that is distributed with SPM software and in
the CANlab Core Tools
Alternative in CANlab tools: which('gray_matter_mask.img')
'noverbose' : Suppress verbose output during image loading
'sample2mask' : Sample images to mask space. Default: Sample mask to
image space, use native image space
Creating class instances
-----------------------------------------------------------------------
You can create an empty object by using:
fmri_dat = fmri_data
- fmri_dat is the object.
- It will be created with a standard brain mask, brainmask.nii
- This image should be placed on your Matlab path
- The space information is stored in fmri_dat.volInfo
- Data is stored in fmri_dat.dat, in a [voxels x images] matrix
- You can replace or append data to the fmri_dat.dat field.
You can create an object by assembling an image_vector object from parts
(entering fields) and converting using fmri_obj = fmri_data(image_vec_obj)
You can create an fmri_data object with extacted image data.
- Let "imgs" be a string array or cell array of image names
- This command creates an object with your (4-D) image data:
- fmri_dat = fmri_data(imgs);
- Images can be zipped (.gz) as well. fmri_data() will unpack them.
- Only values in the standard brain mask, brainmask.nii, will be included.
- This saves memory by reducing the number of voxels saved dramatically.
You can specify any mask you'd like to extract data from.
- Let "maskimagename" be a string array with a mask image name.
- this command creates the object with data saved in the mask:
- fmri_dat = fmri_data(imgs, maskimagename);
- The mask information is saved in fmri_dat.mask
e.g., this extracts data from images within the standard brain mask:
dat = fmri_data(imgs, which('brainmask.nii'));
Defining the space of the extracted data
-----------------------------------------------------------------------
Note: There are two options for defining the space (i.e., coordinates/voxels)
that the data is mapped to.
By default, the mask is resliced to the same space as the first image in the
input image name set (not coregistered; just resliced to the same voxel sizes.
The images are assumed to be in register.)
Reampling to mask space: YOU CAN ALSO map the image data to the space of the mask, by entering
'sample2mask' as in input argument.
For loading images in different spaces together in one object, use the 'sample2mask' option.
Attaching additional data
-----------------------------------------------------------------------
The fmri_data object has a number of fields for appending specific types of data.
- You can replace or append data to the fmri_dat.dat field.
- The fmri_data object will also store predictor data (.X) also outcome data (.Y)
- There are many fields for descriptions, notes, etc., like "dat_descrip" and "source_notes"
- Attach custom descriptions in these fields to document your object.
- The "history" field stores a cell array of strings with the processing
history of the object. Some methods add to this history automatically.
-----------------------------------------------------------------------
Properties and methods
-----------------------------------------------------------------------
Properties are data fields associated with an object.
Type the name of an object (class instance) you create to see its
properties, and a link to its methods (things you can run specifically
with this object type). For example: After creating an fmri_data object
called fmri_dat, as above, type fmri_dat to see its properties.
There are many other methods that you can apply to fmri_data objects to
do different things.
- Try typing methods(fmri_data) for a list.
- You always pass in an fmri_data object as the first argument.
- Methods include utilities for many functions - e.g.,:
- resample_space(fmri_dat) resamples the voxels
- write(fmri_dat) writes an image file to disk (careful not to overwrite by accident!)
- regress(fmri_dat) runs multiple regression
- predict(fmri_dat) runs cross-validated machine learning/prediction algorithms
Key properties and methods (a partial list; type doc fmri_data for more):
-------------------------------------------------------------------------
fmri_data Properties (a partial list; type doc fmri_data for more):
dat - Image data, a [voxels x images] matrix, single-format
fullpath - List of image names loaded into object with full paths
history - History of object processing, for provenance
image_names - List of image names loaded into object, no paths
removed_images - Vector of images that have been removed (saves space; see remove_empty.m, replace_empty.m)
removed_voxels - Vector of empty in-mask voxels that have been removed (saves space; see remove_empty.m, replace_empty.m)
volInfo - Structure with info on brain mask (saves space) and mapping voxels to brain space
fmri_data Methods (a partial list; type doc fmri_data for more):
General:
. descriptives - Get descriptives for an fmri_data or other image_vector object
enforce_variable_types - Re-casts variables in objects into standard data types, which can save
flip - Flips images stored in an object left-to-right
history - Display history for image_vector object
write - Write an image_vector object to hard drive as an Analyze image (uses .fullpath field for image names)
Data extraction:
apply_atlas - Computes the mean value or pattern expression for each reference region specified in an atlas object
apply_mask - Apply a mask image (image filename or fmri_mask_image object) to an image_vector object
apply_parcellation - Computes the mean value or pattern expression for each parcel specified in a data object
extract_gray_white_csf - Extracts mean values (values) and top 5 component scores (components)
extract_roi_averages - This image_vector method a extracts and averages data stored in an fmri_data object
Handling brain space and image selection:
compare_space - Compare spaces of two image_vector objects
get_wh_image - For an image_vector with multiple images (cases, contrasts, etc.), select a subset.
reconstruct_image - Reconstruct a 3-D or 4-D image from image_vector object obj
remove_empty - remove vox: logical vector of custom voxels to remove, VOX x 1
reparse_contiguous - Re-construct list of contiguous voxels in an image based on in-image
replace_empty - Replace empty/missing values in an image data object
resample_space - Resample the images in an fmri_data object (obj) to the space of another
Display and visualization:
display_slices - Creates 3 separate montage views - ax, cor, sagg in a special figure window
histogram - Create a histogram of image values or a series of histograms for each
image_similarity_plot - Associations between images in object and set of 'spatial basis function' images (e.g., 'signatures' or pre-defined maps)
isosurface - Create and visualize an isosurface created from the boundaries in an image object.
montage - Create a montage of an image_vector (or statistic_image or fmri_data)
orthviews - display SPM orthviews for CANlab image_vector (or fmri_data, statistic_image) object
pattern_surf_plot_mip - axial maximum intensity projection pattern surface plot
sagg_slice_movie - Movie of successive differences (sagittal slice)
slices - Create a montage of single-slice results for every image in an image_vector object
surface - Render image data on brain surfaces; options for cutaways and canonical surfaces
wedge_plot_by_atlas - Plot a data object or 'signature' pattern divided into local regions
Data processing and analysis:
ica - Spatial ICA of an fmri_data object
image_math - Perform simple mathematical and boolean operations on image objects (see also plus, minus, power)
mahal - Mahalanobis distance for each image in a set compared to others in the set
mean - Mean across a set of images. Returns a new image_vector object.
preprocess - Preprocesses data in an image_vector (e.g., fmri_data) object; many options for filtering and outlier id
qc_metrics_second_level - Quality metrics for a 2nd-level analysis (set of images from different subjects)
searchlight - Run searchlight multivariate prediction/classification on an image_vector
threshold - Threshold image_vector (or fmri_data or fmri_obj_image) object based on raw threshold values
union - ...and intersection masks for two image_vector objects
-------------------------------------------------------------------------
Examples and help:
-------------------------------------------------------------------------
To list properties and methods for this object, type:
doc fmri_data, methods(fmri_data)
Example 1: Load images (and run a simple analysis)
Load a sample dataset into an fmri_data object (subclass of image_vector)
This loads one of a set of named image collections used in demos/help:
data_obj = load_image_set('emotionreg');
You can load the same images manually, by locating the files, listing
their names in a character array (or 1 x n cell array of strings), and
then passing those into fmri_data:
data_obj = fmri_data(which('Wager_2008_emo_reg_vs_look_neg_contrast_images.nii.gz'));
filedir = what(fullfile('CanlabCore', 'Sample_datasets', 'Wager_et_al_2008_Neuron_EmotionReg'));
image_names = filenames(fullfile(filedir.path, '*img'));
data_obj = fmri_data(image_names);
Now you can interact with the object. Try, e.g.,:
methods(data_obj) % List methods for object type
descriptives(data_obj); % Print summary of descriptive statistics for the dataset
plot(data_obj) % Custom fmri_data specific plots
t = ttest(data_obj); % Perform a voxel-wise one-sample t-test across images
t = threshold(t, .005, 'unc', 'k', 10); % Re-threshold with extent threshold of 10 contiguous voxels
r = region(t); % Turn t-map into a region object with one element per contig region
Example 2: Extract data averaged over regions of interest:
First run Example 1. Now you have a thresholded t-statistic map.
Extract averages (across voxels) for each subject in each contiguous
region by typing:
r = extract_roi_averages(data_obj, t);
This returns r, which is another object--a "region"-class object.
Region objects contain vectors, one element per pre-defined region in the
image (in this case, significant blobs from our analysis).
Each element contains info that describes the region, including the voxels included,
and average and voxel-by-voxel data if extracted from images and attached.
Type "doc region" for more info.
r has properties that hold multiple types of data:
- .dat holds generic extracted data
- .all_data holds voxel-by-voxel extracted data
For more examples and walkthroughs, see the CANlab_help_examples
repository at https://github.com/canlab/CANlab_help_examples
Some example tutorials:
canlab_help_1_installing_tools
canlab_help_2_load_a_sample_dataset
canlab_help_3_voxelwise_t_test_walkthrough
canlab_help_4_write_data_to_image_file_format
canlab_help_5_regression_walkthrough
Reference page in Doc Center
doc fmri_data
Section 5: Explore methods
Objects have methods associated with them, or things you can do with them. To see a list of methods for the fmri_data object, type:
methods(fmri_data)
Methods for class fmri_data: apply_atlas plus apply_mask power apply_parcellation prctile canlab_connectivity_preproc predict cat predict_test_suite check_image_filenames preprocess compare_space qc_metrics_second_level create read_from_file descriptives rebuild_volinfo_from_dat display_slices reconstruct_image enforce_variable_types regress expand_into_atlas_subregions remove_empty extract_gray_white_csf reparse_contiguous extract_measures_batch replace_empty extract_roi_averages resample_space flip resample_time fmri_data rescale get_wh_image riverplot histogram rsa_regression history sagg_slice_movie horzcat saveplots hrf_fit searchlight ica select_voxels_by_value image_math signtest image_similarity_plot sim_data image_similarity_plot_bucknermaps slices interpolate split isempty surface isosurface test_generalizability mahal threshold mean trim_mask minus ttest montage ttest_table_and_lateralization_test neurosynth_feature_labels union orthviews wedge_plot_by_atlas pattern_surf_plot_mip windsorize plot write plot_current_orthviews_coord
You can also use the name of an instance of an object, i.e., a variable in the workspace. methods(data_obj) produces the same list. In addition, you can get help on any of the object's methods by typing: object class name_dot_method name. e.g.,
help fmri_data.mean % Note: There may be trouble accessing these help files in Matlab R2019b. % Works in 2018 and earlier. But maybe this is not a general problem...give % it a try.
Mean across a set of images. Returns a new image_vector object.
Creates an image_vector object with mean values for each voxel (cols)
across images (rows) of an image_vector (e.g., fmri_data) object.
:Usage:
::
function [m, imagemeans, voxelmeans] = mean(obj, [optional args])
m is an image_vector object whose data contains the mean values.
- Average available valid data in each voxel. Some images may have
missing data for some voxels.
:Optional Inputs:
- 'write', followed by file name
- 'path', followed by location for file (default = current directory)
- 'orthviews' -> show orthviews for this image, same as orthviews(m)
- 'histogram' -> show histogram for this image, same as histogram(m)
- 'plot' -> do both
:Examples:
::
% If sdat is an fmri_data object with multiple images,
m = mean(sdat, 'plot', 'write', anatmeanname, 'path', maskdir);
Help for fmri_data/mean is inherited from superclass IMAGE_VECTOR
Section 6: Explore on your own
1. Try to run a couple of other methods on your fmri_data object. Some require additional inputs, but many do not. Look at the help for more info. What do you see? Can you return any meaningful output, and if so, what did the method do?
% That's it for this section!!
Explore More: CANlab Toolboxes
Tutorials, overview, and help: https://canlab.github.io
Toolboxes and image repositories on github: https://github.com/canlab
| CANlab Core Tools | https://github.com/canlab/CanlabCore | CANlab Neuroimaging_Pattern_Masks repository | https://github.com/canlab/Neuroimaging_Pattern_Masks | CANlab_help_examples | https://github.com/canlab/CANlab_help_examples | M3 Multilevel mediation toolbox | https://github.com/canlab/MediationToolbox | M3 CANlab robust regression toolbox | https://github.com/canlab/RobustToolbox | M3 MKDA coordinate-based meta-analysis toolbox | https://github.com/canlab/Canlab_MKDA_MetaAnalysis |
Here are some other useful CANlab-associated resources:
| Paradigms_Public - CANlab experimental paradigms | https://github.com/canlab/Paradigms_Public | FMRI_simulations - brain movies, effect size/power | https://github.com/canlab/FMRI_simulations | CANlab_data_public - Published datasets | https://github.com/canlab/CANlab_data_public | M3 Neurosynth: Tal Yarkoni | https://github.com/neurosynth/neurosynth | M3 DCC - Martin Lindquist's dynamic correlation tbx | https://github.com/canlab/Lindquist_Dynamic_Correlation | M3 CanlabScripts - in-lab Matlab/python/bash | https://github.com/canlab/CanlabScripts |
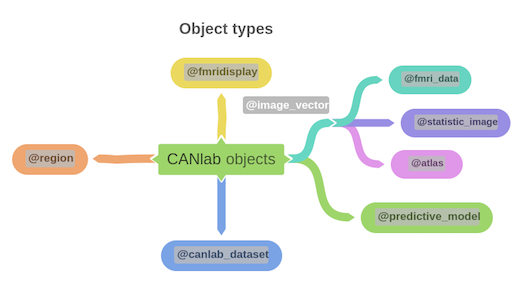
Object-oriented, interactive approach The core basis for interacting with CANlab tools is through object-oriented framework. A simple set of neuroimaging data-specific objects (or classes) allows you to perform interactive analysis using simple commands (called methods) that operate on objects.
Map of core object classes: