Object-oriented analysis
The CANlab imaging analysis tools consist of a set of linked Github repositories.
They enable interactive data analysis for fMRI and other types of neuroimaging data using objects with simple methods (or commands) customized for neuroimaging data analysis.
Interactive analysis: Philosophy and principles
Interactive analysis using objects has a number of advantages. See the main CANlab intro page for more on the philosophy behind this approach.
Locations of walkthroughs and batch scripts
install. How to install the tools.
Other step-by-step analysis walkthroughs are in the CANlab_help_examples repository, in the example_help_files folder.
These are executable Matlab files (.m files) that can be run. They can also be published to HTML files (see canlab_help_publish.m). A set of HTML files with figures and printouts of these help examples is in the published_html folder.
A system of batch scripts is in the CANlab_help_examples repository, in the Second_level_analysis_template_scripts folder. See batch workflow for a walkthrough of the batch scripts.
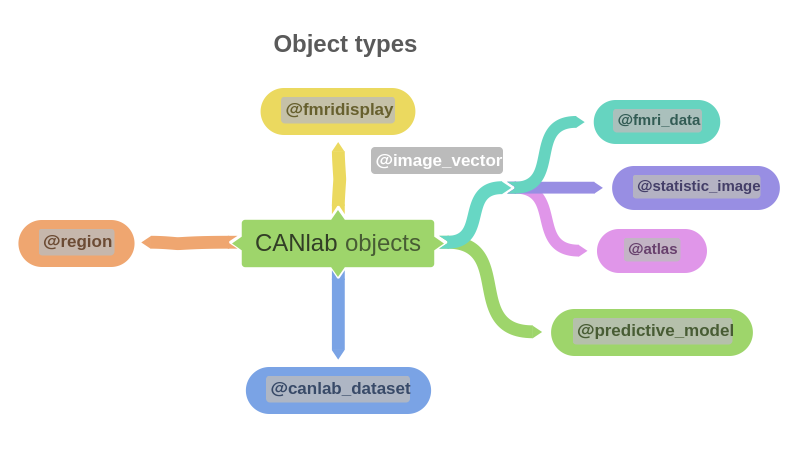
types of objects
| Object | Description |
|---|---|
| fmri_data | Stores neuroimaging datasets; operate on datasets by calling object methods |
| statistic_image | Stores statistic images with stats and P-values |
| atlas | Stores atlas data with probability maps and integers for unique regions |
| region | Stores data grouped by region/network, treating region as a unit of analysis |
| fmridisplay | Container for handles for brain slices and surfaces, allowing visualization |
fmri_data object features and philosophy
- Flat format: Store image data in a flat (2-d), space-efficient voxels x images matrix
- Save space: Tools to remove out-of-image voxels and empty (zero/NaN) voxels, store data in single-precision format
- Analysis-friendly: 2-d matrices can be read and analyzed in multiple packages/algorithms
- Meta-data included to convert back to 3-d image volume space, with easy tools (methods) to reconstruct and visualize images Built-in resampling makes it easy to compare/combine datasets with different voxel sizes and image bounding boxes. Reduces overhead for statisticians/data scientists unfamiliar with neuroimaging to apply their algorithms.
- Multiple images can be stored in a single object
- Methods have short, intuitive names, and perform high-level functions specialized for neuroimaging. e.g., some example methods are:
- Visualization (plot, orthviews, surface, montage, histogram, isosurface methods)
- Image manipulation (apply_mask, get_wh_image, resample_space, compare_space, flip, threshold methods)
- Data extraction (apply_atlas, apply_parcellation, extract_gray_white_csf, extract_roi_averages)
- Analysis (ica, mahal, image_math, and many more in the fmri_data subclass)
- Provenance: Ability to track and update history of changes to objects
- Documentation: In Matlab, type
doc object_classname (e.g.,doc fmri_data) for properties, methods, and examples.
flowchart for a simple Analysis
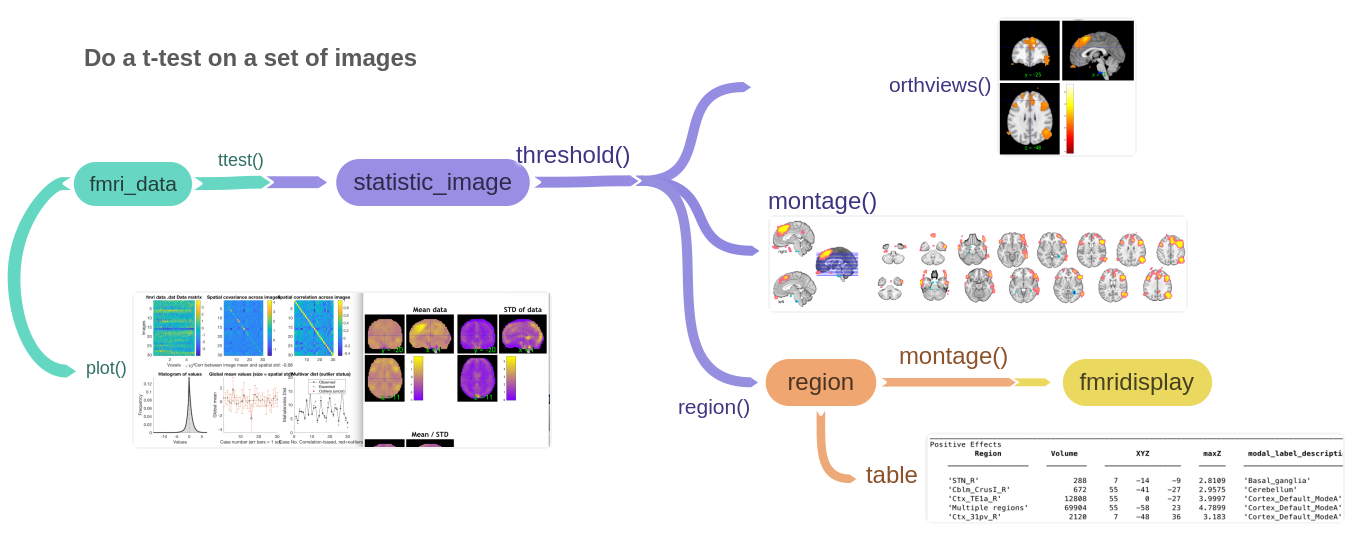
The diagram below shows a flowchart for a simple group analysis, starting by loading a set of images (.nii or .img, one per subject) into an fmri_data object. A few simple commands can perform a t-test (or other analysis), threshold the resulting statistic map (a statistic_image object), and display interactive views, slice montages, tables with automatically labeled regions, and more.